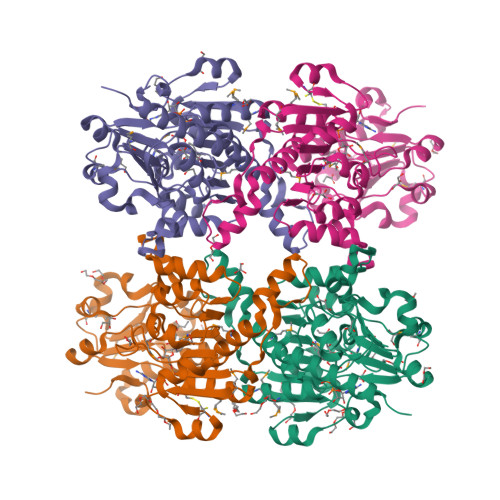
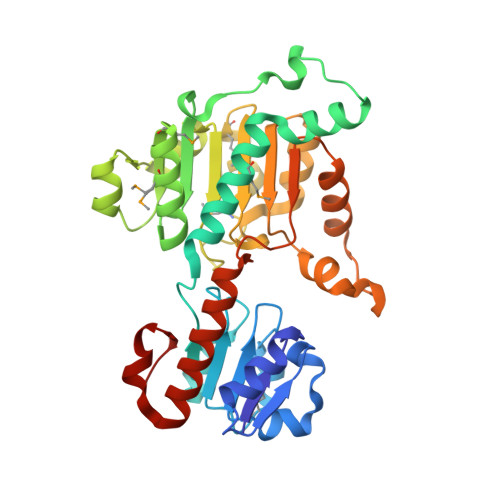
Structure of D-lactate dehydrogenase from Aquifex aeolicus complexed with NAD(+) and lactic acid (or pyruvate).
Antonyuk, S.V., Strange, R.W., Ellis, M.J., Bessho, Y., Kuramitsu, S., Inoue, Y., Yokoyama, S., Hasnain, S.S.(2009) Acta Crystallogr Sect F Struct Biol Cryst Commun 65: 1209-1213
- PubMed: 20054113
- DOI: https://doi.org/10.1107/S1744309109044935
- Primary Citation of Related Structures:
3KB6 - PubMed Abstract:
The crystal structure of D-lactate dehydrogenase from Aquifex aeolicus (aq_727) was determined to 2.12 A resolution in space group P2(1)2(1)2(1), with unit-cell parameters a = 90.94, b = 94.43, c = 188.85 A. The structure was solved by molecular replacement using the coenzyme-binding domain of Lactobacillus helveticus D-lactate dehydrogenase and contained two homodimers in the asymmetric unit. Each subunit of the homodimer was found to be in a ;closed' conformation with the NADH cofactor bound to the coenzyme-binding domain and with a lactate (or pyruvate) molecule bound at the interdomain active-site cleft.
Organizational Affiliation:
Molecular Biophysics Group, School of Biological Sciences, University of Liverpool, Crown Street, Liverpool L69 7ZB, England.