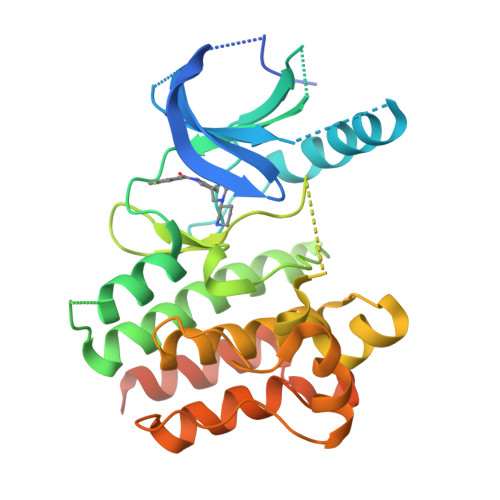
Novel and selective spiroindoline-based inhibitors of sky kinase.
Powell, N.A., Kohrt, J.T., Filipski, K.J., Kaufman, M., Sheehan, D., Edmunds, J.E., Delaney, A., Wang, Y., Bourbonais, F., Lee, D.Y., Schwende, F., Sun, F., McConnell, P., Catana, C., Chen, H., Ohren, J., Perrin, L.A.(2012) Bioorg Med Chem Lett 22: 190-193
- PubMed: 22119469
- DOI: https://doi.org/10.1016/j.bmcl.2011.11.036
- Primary Citation of Related Structures:
3QUP - PubMed Abstract:
We report the discovery of a novel series of spiroindoline-based inhibitors of Sky kinase that bind in the ATP-binding site and exhibit high levels of kinome selectivity through filling the Ala571-subpocket. These inhibitors exhibit moderate oral bioavailability in the rat due to low absorption across the gut wall.
Organizational Affiliation:
Pfizer Global Research & Development, Michigan Laboratories, Ann Arbor, MI 48105, USA. napowell@comcast.net