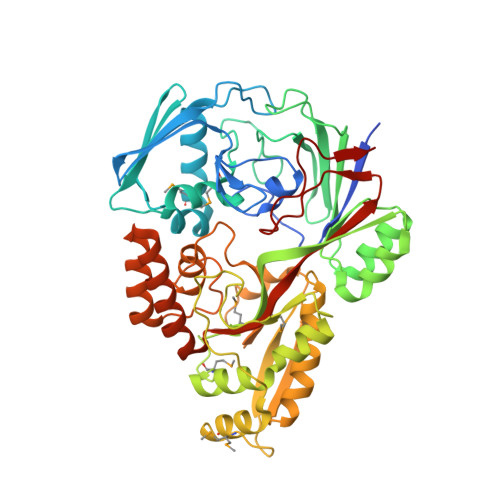
1.5 Angstrom Crystal Structure of the Complex of Ligand Binding Component of ABC-type Import System from Staphylococcus aureus with Nickel and two Histidines.
Minasov, G., Halavaty, A., Shuvalova, L., Dubrovska, I., Winsor, J., Kiryukhina, O., Falugi, F., Bottomley, M., Bagnoli, F., Grandi, G., Anderson, W.F.To be published.