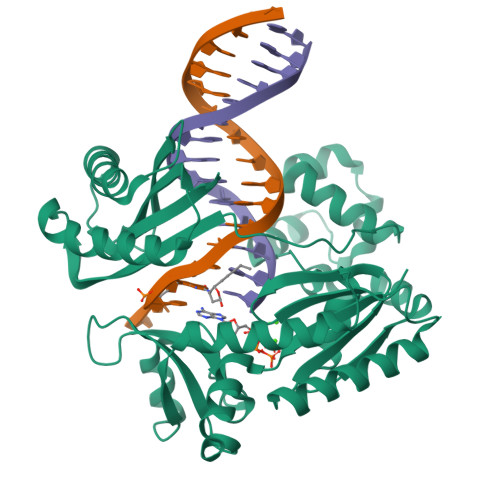
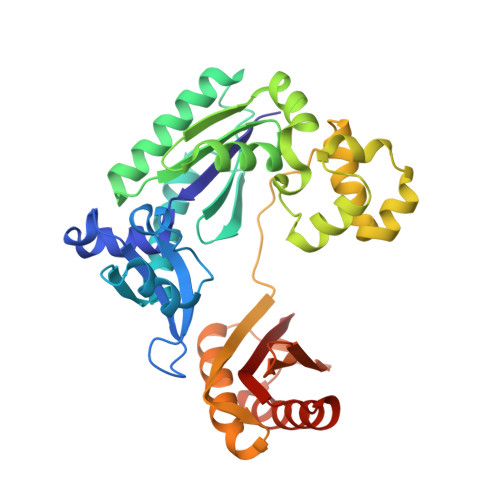
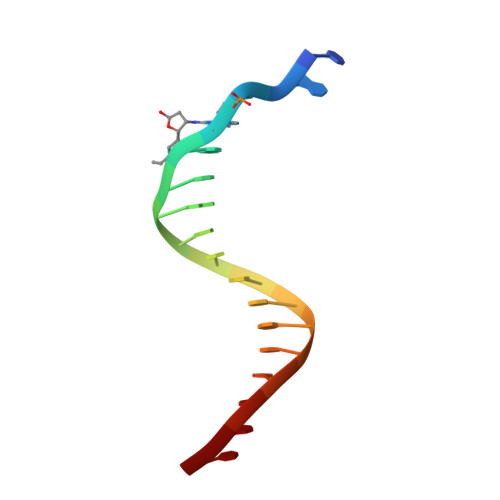
Replication bypass of the trans-4-Hydroxynonenal-derived (6S,8R,11S)-1,N(2)-deoxyguanosine DNA adduct by the sulfolobus solfataricus DNA polymerase IV.
Banerjee, S., Christov, P.P., Kozekova, A., Rizzo, C.J., Egli, M., Stone, M.P.(2012) Chem Res Toxicol 25: 422-435
- PubMed: 22313351
- DOI: https://doi.org/10.1021/tx200460j
- Primary Citation of Related Structures:
3T5H, 3T5J, 3T5K, 3T5L - PubMed Abstract:
trans-4-Hydroxynonenal (HNE) is the major peroxidation product of ω-6 polyunsaturated fatty acids in vivo. Michael addition of the N(2)-amino group of dGuo to HNE followed by ring closure of N1 onto the aldehyde results in four diastereomeric 1,N(2)-dGuo (1,N(2)-HNE-dGuo) adducts. The (6S,8R,11S)-HNE-1,N(2)-dGuo adduct was incorporated into the 18-mer templates 5'-d(TCATXGAATCCTTCCCCC)-3' and d(TCACXGAATCCTTCCCCC)-3', where X = (6S,8R,11S)-HNE-1,N(2)-dGuo adduct. These differed in the identity of the template 5'-neighbor base, which was either Thy or Cyt, respectively. Each of these templates was annealed with either a 13-mer primer 5'-d(GGGGGAAGGATTC)-3' or a 14-mer primer 5'-d(GGGGGAAGGATTCC)-3'. The addition of dNTPs to the 13-mer primer allowed analysis of dNTP insertion opposite to the (6S,8R,11S)-HNE-1,N(2)-dGuo adduct, whereas the 14-mer primer allowed analysis of dNTP extension past a primed (6S,8R,11S)-HNE-1,N(2)-dGuo:dCyd pair. The Sulfolobus solfataricus P2 DNA polymerase IV (Dpo4) belongs to the Y-family of error-prone polymerases. Replication bypass studies in vitro reveal that this polymerase inserted dNTPs opposite the (6S,8R,11S)-HNE-1,N(2)-dGuo adduct in a sequence-specific manner. If the template 5'-neighbor base was dCyt, the polymerase inserted primarily dGTP, whereas if the template 5'-neighbor base was dThy, the polymerase inserted primarily dATP. The latter event would predict low levels of Gua → Thy mutations during replication bypass when the template 5'-neighbor base is dThy. When presented with a primed (6S,8R,11S)-HNE-1,N(2)-dGuo:dCyd pair, the polymerase conducted full-length primer extension. Structures for ternary (Dpo4-DNA-dNTP) complexes with all four template-primers were obtained. For the 18-mer:13-mer template-primers in which the polymerase was confronted with the (6S,8R,11S)-HNE-1,N(2)-dGuo adduct, the (6S,8R,11S)-1,N(2)-dGuo lesion remained in the ring-closed conformation at the active site. The incoming dNTP, either dGTP or dATP, was positioned with Watson-Crick pairing opposite the template 5'-neighbor base, dCyt or dThy, respectively. In contrast, for the 18-mer:14-mer template-primers with a primed (6S,8R,11S)-HNE-1,N(2)-dGuo:dCyd pair, ring opening of the adduct to the corresponding N(2)-dGuo aldehyde species occurred. This allowed Watson-Crick base pairing at the (6S,8R,11S)-HNE-1,N(2)-dGuo:dCyd pair.
Organizational Affiliation:
Departments of Chemistry and Biochemistry, Center in Molecular Toxicology, Vanderbilt Institute of Chemical Biology and the Vanderbilt-Ingram Cancer Center, Vanderbilt University , Nashville, Tennessee 37235, United States.