Structural studies of hydroxylamine oxidoreductase reveal a unique heme cofactor and a previously unidentified interaction partner.
Cedervall, P.E., Hooper, A.B., Wilmot, C.M.(2013) Biochemistry 52: 6211-6218
- PubMed: 23952581
- DOI: https://doi.org/10.1021/bi400960w
- Primary Citation of Related Structures:
4FAS - PubMed Abstract:
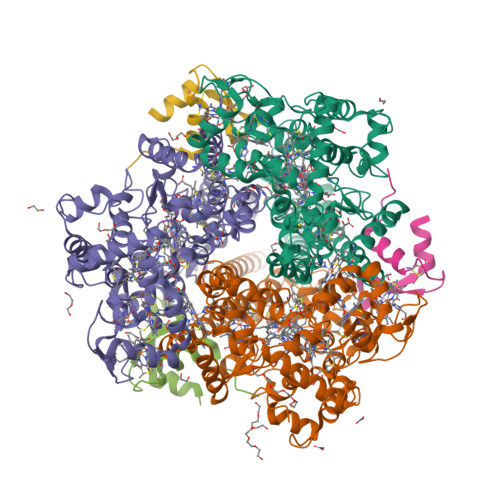
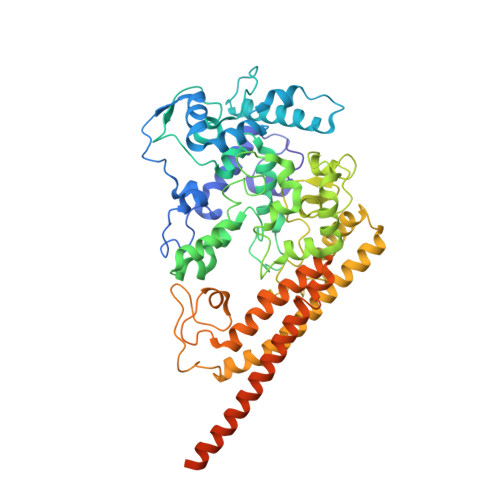
Hydroxylamine oxidoreductase (HAO) is a 24-heme homotrimeric enzyme that catalyzes the conversion of hydroxylamine to nitrite in nitrifying bacteria: a key reaction in the nitrogen cycle. One heme in each HAO monomer is a highly unusual heme P460 that is the site of catalysis. This was proposed to be a c-type heme that contained an additional porphyrin-tyrosine cross-link. Here, we report the crystal structure of HAO from Nitrosomonas europaea to 2.1 Å resolution that defines a different model compatible with the crystallographic and biochemical data. The structure reveals that heme P460 contains two covalent cross-links between the porphyrin and a Tyr residue. In addition, the enzyme was purified from source, and an unknown physiological HAO binding partner was present within the crystal (annotated in the genome as hypothetical protein NE1300). NE1300 may play a structural role in the ternary complex with cytochrome c554, the physiological electron acceptor of HAO.
Organizational Affiliation:
Department of Biochemistry, Molecular Biology, and Biophysics, University of Minnesota , Minneapolis, Minnesota 55455, United States.