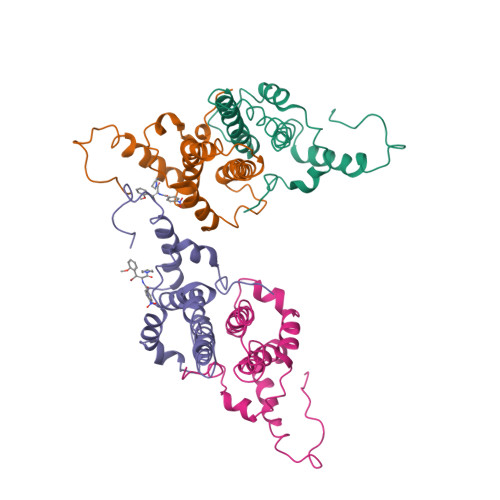
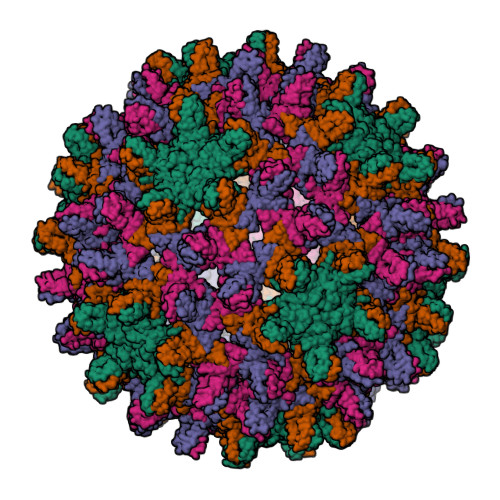
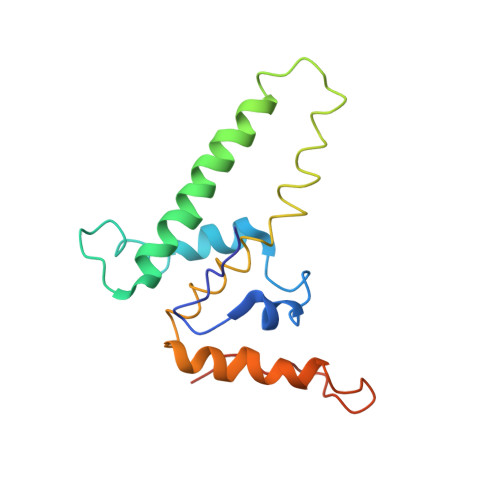
Assembly-directed antivirals differentially bind quasiequivalent pockets to modify hepatitis B virus capsid tertiary and quaternary structure.
Katen, S.P., Tan, Z., Chirapu, S.R., Finn, M.G., Zlotnick, A.(2013) Structure 21: 1406-1416
- PubMed: 23871485
- DOI: https://doi.org/10.1016/j.str.2013.06.013
- Primary Citation of Related Structures:
4G93 - PubMed Abstract:
Hepatitis B virus (HBV) is a major cause of liver disease. Assembly of the HBV capsid is a critical step in virus production and an attractive target for new antiviral therapies. We determined the structure of HBV capsid in complex with AT-130, a member of the phenylpropenamide family of assembly effectors. AT-130 causes tertiary and quaternary structural changes but does not disrupt capsid structure. AT-130 binds a hydrophobic pocket that also accommodates the previously characterized heteroaryldihydropyrimidine compounds but favors a unique quasiequivalent location on the capsid surface. Thus, this pocket is a promiscuous drug-binding site and a likely target for different assembly effectors with a broad range of mechanisms of activity. That AT-130 successfully decreases virus production by increasing capsid assembly rate without disrupting capsid structure delineates a paradigm in antiviral design, that disrupting reaction timing is a viable strategy for assembly effectors of HBV and other viruses.
Organizational Affiliation:
Department of Molecular and Cellular Biochemistry, Indiana University, Bloomington, IN 47405, USA.