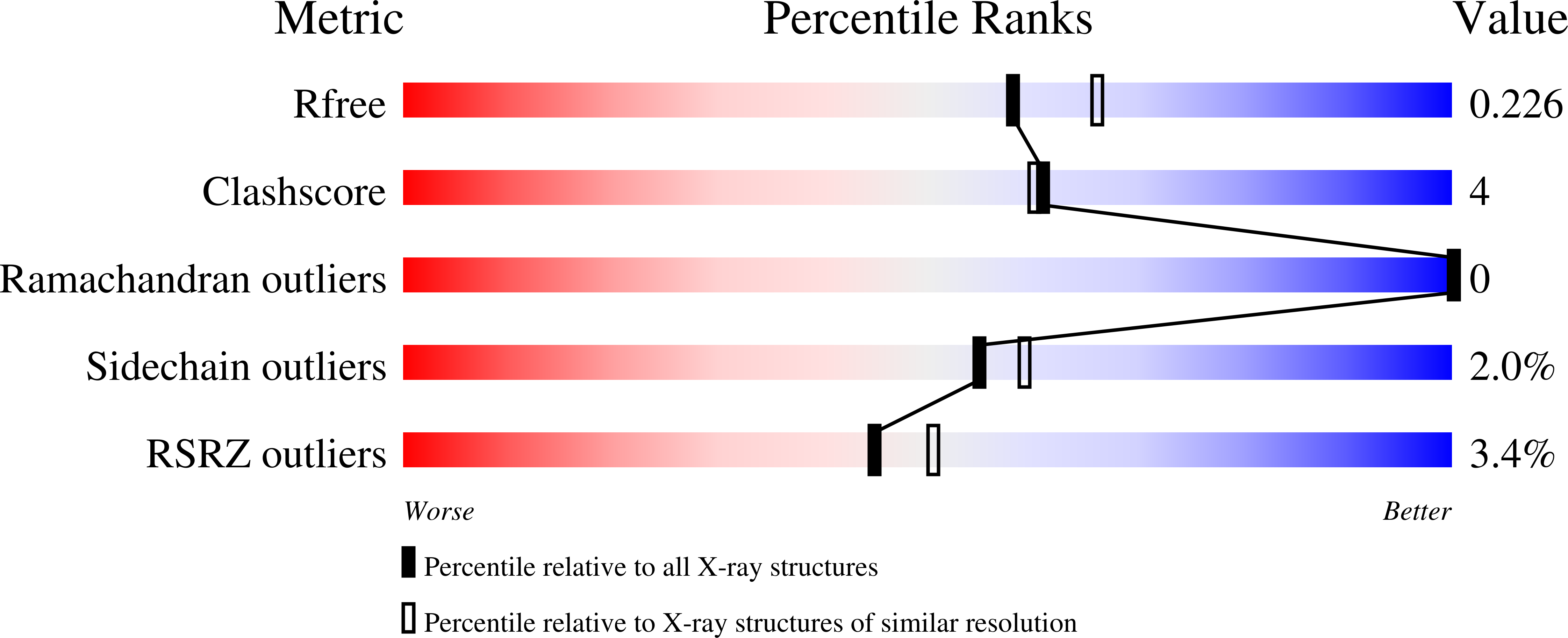
Tyr26 phosphorylation of PGAM1 provides a metabolic advantage to tumours by stabilizing the active conformation.
Hitosugi, T., Zhou, L., Fan, J., Elf, S., Zhang, L., Xie, J., Wang, Y., Gu, T.L., Aleckovic, M., Leroy, G., Kang, Y., Kang, H.B., Seo, J.H., Shan, C., Jin, P., Gong, W., Lonial, S., Arellano, M.L., Khoury, H.J., Chen, G.Z., Shin, D.M., Khuri, F.R., Boggon, T.J., Kang, S., He, C., Chen, J.(2013) Nat Commun 4: 1790-1790
- PubMed: 23653202
- DOI: https://doi.org/10.1038/ncomms2759
- Primary Citation of Related Structures:
4GPI, 4GPZ - PubMed Abstract:
How oncogenic signalling coordinates glycolysis and anabolic biosynthesis in cancer cells remains unclear. We recently reported that the glycolytic enzyme phosphoglycerate mutase 1 (PGAM1) regulates anabolic biosynthesis by controlling intracellular levels of its substrate 3-phosphoglycerate and product 2-phosphoglycerate. Here we report a novel mechanism in which Y26 phosphorylation enhances PGAM1 activation through release of inhibitory E19 that blocks the active site, stabilising cofactor 2,3-bisphosphoglycerate binding and H11 phosphorylation. We also report the crystal structure of H11-phosphorylated PGAM1 and find that phospho-H11 activates PGAM1 at least in part by promoting substrate 3-phosphoglycerate binding. Moreover, Y26 phosphorylation of PGAM1 is common in human cancer cells and contributes to regulation of 3-phosphoglycerate and 2-phosphoglycerate levels, promoting cancer cell proliferation and tumour growth. As PGAM1 is a negative transcriptional target of TP53, and is therefore commonly upregulated in human cancers, these findings suggest that Y26 phosphorylation represents an additional acute mechanism underlying phosphoglycerate mutase 1 upregulation.
Organizational Affiliation:
Department of Haematology and Medical Oncology, Winship Cancer Institute of Emory, Emory University School of Medicine, Atlanta, Georgia 30322, USA.