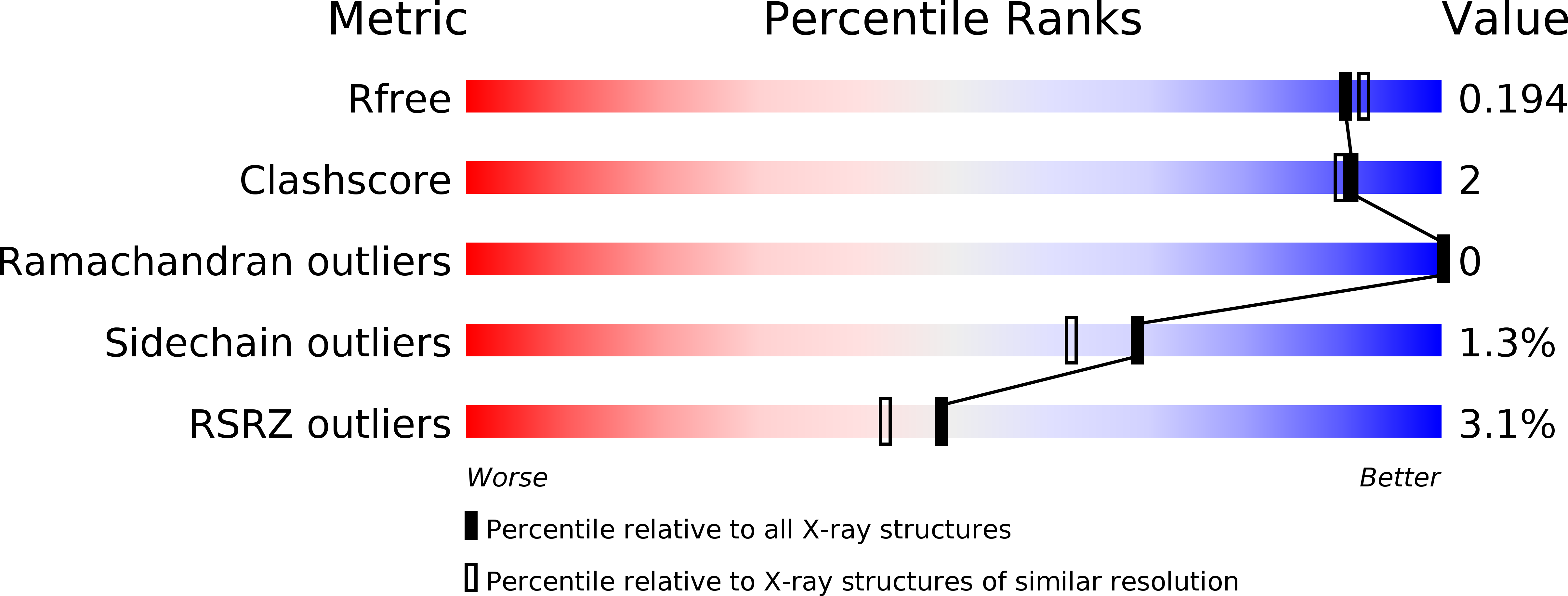
Crystal Structure and Site-Directed Mutagenesis Analyses of Haloalkane Dehalogenase LinB from Sphingobium sp. Strain MI1205.
Okai, M., Ohtsuka, J., Imai, L.F., Mase, T., Moriuchi, R., Tsuda, M., Nagata, K., Nagata, Y., Tanokura, M.(2013) J Bacteriol 195: 2642-2651
- PubMed: 23564170
- DOI: https://doi.org/10.1128/JB.02020-12
- Primary Citation of Related Structures:
4H77, 4H7D, 4H7E, 4H7F, 4H7H, 4H7I, 4H7J, 4H7K - PubMed Abstract:
The enzymes LinB(UT) and LinB(MI) (LinB from Sphingobium japonicum UT26 and Sphingobium sp. MI1205, respectively) catalyze the hydrolytic dechlorination of β-hexachlorocyclohexane (β-HCH) and yield different products, 2,3,4,5,6-pentachlorocyclohexanol (PCHL) and 2,3,5,6-tetrachlorocyclohexane-1,4-diol (TCDL), respectively, despite their 98% identity in amino acid sequence. To reveal the structural basis of their different enzymatic properties, we performed site-directed mutagenesis and X-ray crystallographic studies of LinB(MI) and its seven point mutants. The mutation analysis revealed that the seven amino acid residues uniquely found in LinB(MI) were categorized into three groups based on the efficiency of the first-step (from β-HCH to PCHL) and second-step (from PCHL to TCDL) conversions. Crystal structure analyses of wild-type LinB(MI) and its seven point mutants indicated how each mutated residue contributed to the first- and second-step conversions by LinB(MI). The dynamics simulation analyses of wild-type LinB(MI) and LinB(UT) revealed that the entrance of the substrate access tunnel of LinB(UT) was more flexible than that of LinB(MI), which could lead to the different efficiencies of dehalogenation activity between these dehalogenases.
Organizational Affiliation:
Department of Applied Biological Chemistry, Graduate School of Agricultural and Life Sciences, University of Tokyo, Tokyo, Japan.