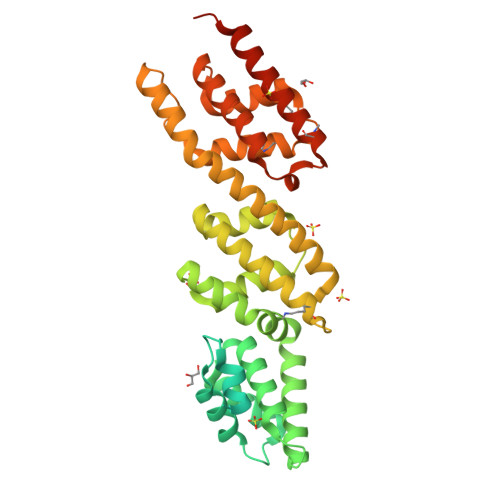
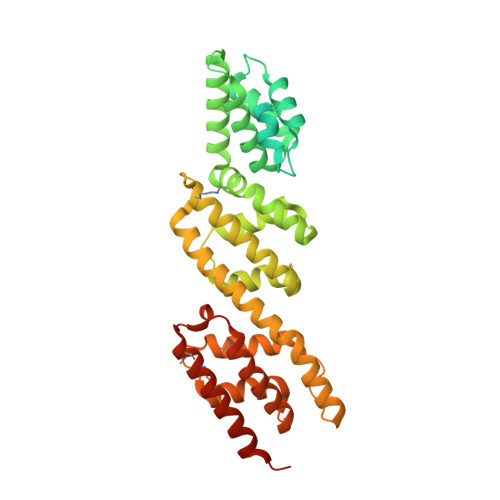
Biochemical and Structural Insights into Microtubule Perturbation by CopN from Chlamydia pneumoniae.
Nawrotek, A., Guimaraes, B.G., Velours, C., Subtil, A., Knossow, M., Gigant, B.(2014) J Biological Chem 289: 25199-25210
- PubMed: 25056950
- DOI: https://doi.org/10.1074/jbc.M114.568436
- Primary Citation of Related Structures:
4P3Z, 4P40 - PubMed Abstract:
Although the actin network is commonly hijacked by pathogens, there are few reports of parasites targeting microtubules. The proposed member of the LcrE protein family from some Chlamydia species (e.g. pCopN from C. pneumoniae) binds tubulin and inhibits microtubule assembly in vitro. From the pCopN structure and its similarity with that of MxiC from Shigella, we definitively confirm CopN as the Chlamydia homolog of the LcrE family of bacterial proteins involved in the regulation of type III secretion. We have also investigated the molecular basis for the pCopN effect on microtubules. We show that pCopN delays microtubule nucleation and acts as a pure tubulin-sequestering protein at steady state. It targets the β subunit interface involved in the tubulin longitudinal self-association in a way that inhibits nucleotide exchange. pCopN contains three repetitions of a helical motif flanked by disordered N- and C-terminal extensions. We have identified the pCopN minimal tubulin-binding region within the second and third repeats. Together with the intriguing observation that C. trachomatis CopN does not bind tubulin, our data support the notion that, in addition to the shared function of type III secretion regulation, these proteins have evolved different functions in the host cytosol. Our results provide a mechanistic framework for understanding the C. pneumoniae CopN-specific inhibition of microtubule assembly.
Organizational Affiliation:
From the Laboratoire d'Enzymologie et Biochimie Structurales (LEBS), Centre de Recherche de Gif, CNRS, 91198 Gif sur Yvette, France.