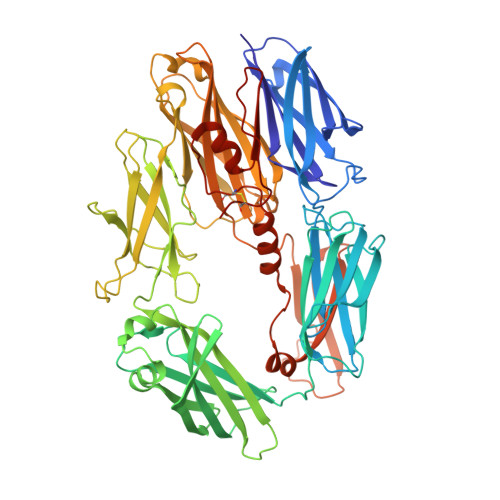
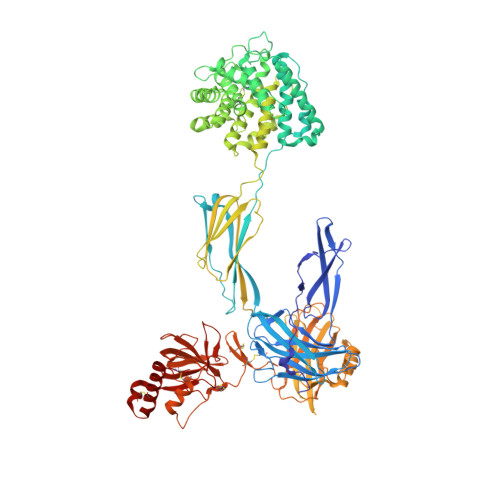
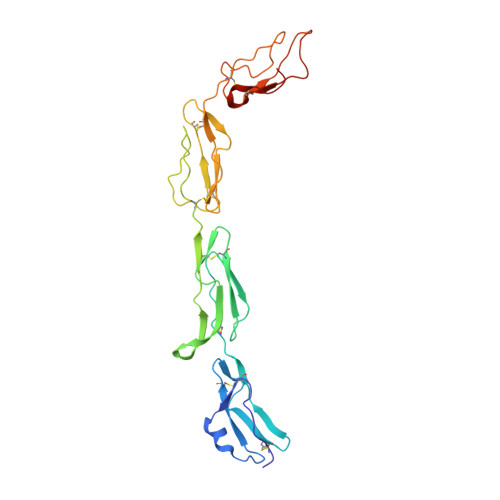
Regulators of Complement Activity Mediate Inhibitory Mechanisms Through a Common C3B-Binding Mode.
Forneris, F., Wu, J., Xue, X., Ricklin, D., Lin, Z., Sfyroera, G., Tzekou, A., Volokhina, E., Granneman, J.C., Hauhart, R., Bertram, P., Liszewski, M.K., Atkinson, J.P., Lambris, J.D., Gros, P.(2016) EMBO J 35: 1133
- PubMed: 27013439
- DOI: https://doi.org/10.15252/embj.201593673
- Primary Citation of Related Structures:
5FO7, 5FO8, 5FO9, 5FOA, 5FOB - PubMed Abstract:
Regulators of complement activation (RCA) inhibit complement-induced immune responses on healthy host tissues. We present crystal structures of human RCA (MCP, DAF, and CR1) and a smallpox virus homolog (SPICE) bound to complement component C3b. Our structural data reveal that up to four consecutive homologous CCP domains (i-iv), responsible for inhibition, bind in the same orientation and extended arrangement at a shared binding platform on C3b. Large sequence variations in CCP domains explain the diverse C3b-binding patterns, with limited or no contribution of some individual domains, while all regulators show extensive contacts with C3b for the domains at the third site. A variation of ~100° rotation around the longitudinal axis is observed for domains binding at the fourth site on C3b, without affecting the overall binding mode. The data suggest a common evolutionary origin for both inhibitory mechanisms, called decay acceleration and cofactor activity, with variable C3b binding through domains at sites ii, iii, and iv, and provide a framework for understanding RCA disease-related mutations and immune evasion.
Organizational Affiliation:
Crystal and Structural Chemistry, Bijvoet Center for Biomolecular Research, Department of Chemistry, Faculty of Science Utrecht University, Utrecht, The Netherlands.