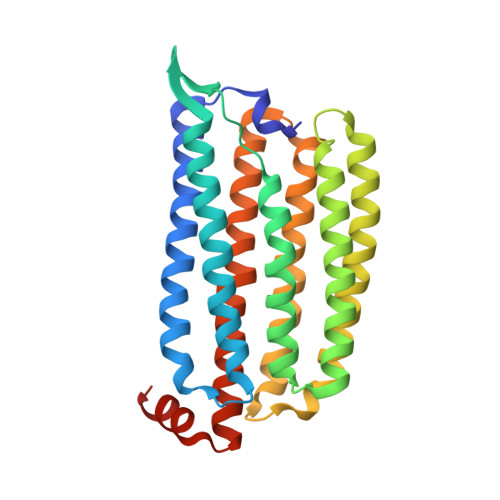
Crystal Structure and Functional Characterization of a Light-Driven Chloride Pump Having an Ntq Motif.
Kim, K., Kwon, S., Jun, S., Cha, J.S., Kim, H., Lee, W., Kim, J.F., Cho, H.(2016) Nat Commun 7: 12677
- PubMed: 27554809
- DOI: https://doi.org/10.1038/ncomms12677
- Primary Citation of Related Structures:
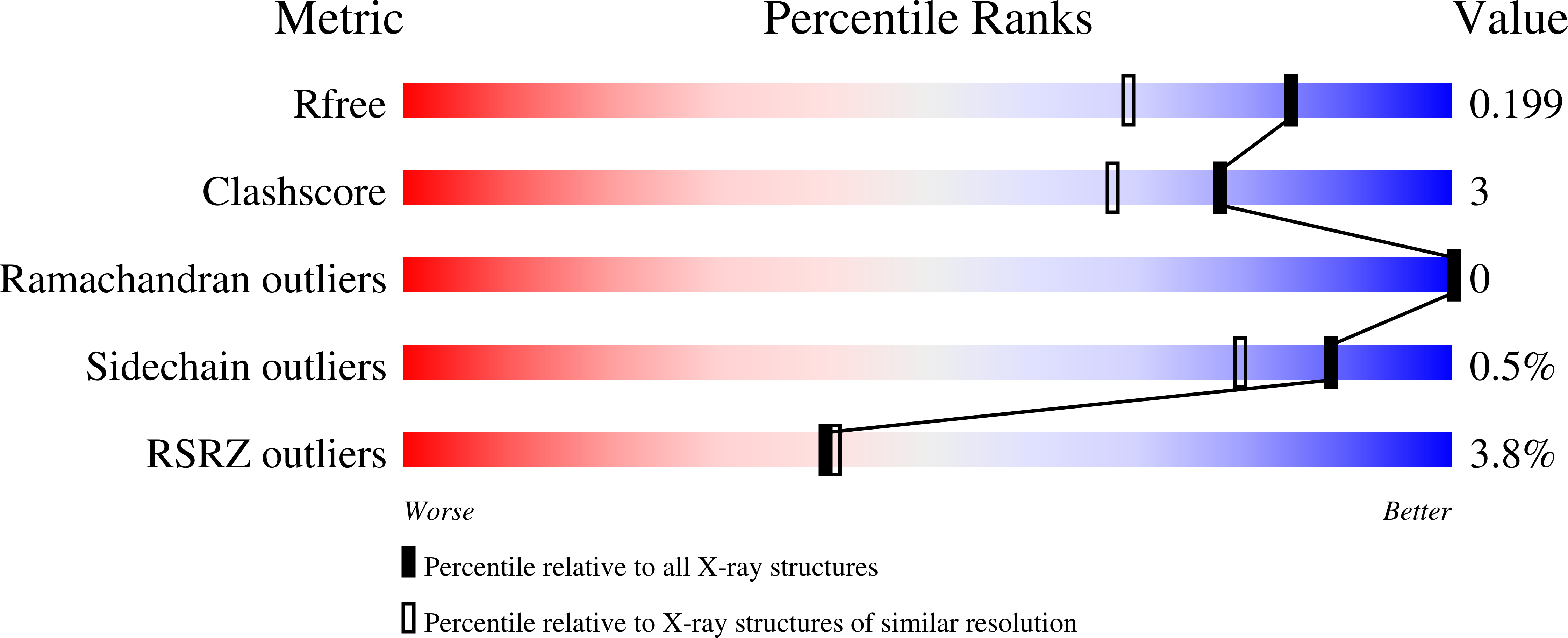
5G28, 5G2A, 5G2C, 5G2D, 5G54 - PubMed Abstract:
A novel light-driven chloride-pumping rhodopsin (ClR) containing an 'NTQ motif' in its putative ion conduction pathway has been discovered and functionally characterized in a genomic analysis study of a marine bacterium. Here we report the crystal structure of ClR from the flavobacterium Nonlabens marinus S1-08(T) determined under two conditions at 2.0 and 1.56 Å resolutions. The structures reveal two chloride-binding sites, one around the protonated Schiff base and the other on a cytoplasmic loop. We identify a '3 omega motif' formed by three non-consecutive aromatic amino acids that is correlated with the B-C loop orientation. Detailed ClR structural analyses with functional studies in E. coli reveal the chloride ion transduction pathway. Our results help understand the molecular mechanism and physiological role of ClR and provide a structural basis for optogenetic applications.
Organizational Affiliation:
Department of Systems Biology and Division of Life Sciences, Yonsei University, 50 Yonsei-ro, Seoul 03722, Republic of Korea.