Structure of protein O-mannose kinase reveals a unique active site architecture
Zhu, Q., Venzke, D., Walimbe, A.S., Anderson, M.E., Fu, Q., Kinch, L.N., Wang, W., Chen, X., Grishin, N.V., Huang, N., Yu, L., Dixon, J.E., Campbell, K.P., Xiao, J.(2016) Elife 5
- PubMed: 27879205
- DOI: https://doi.org/10.7554/eLife.22238
- Primary Citation of Related Structures:
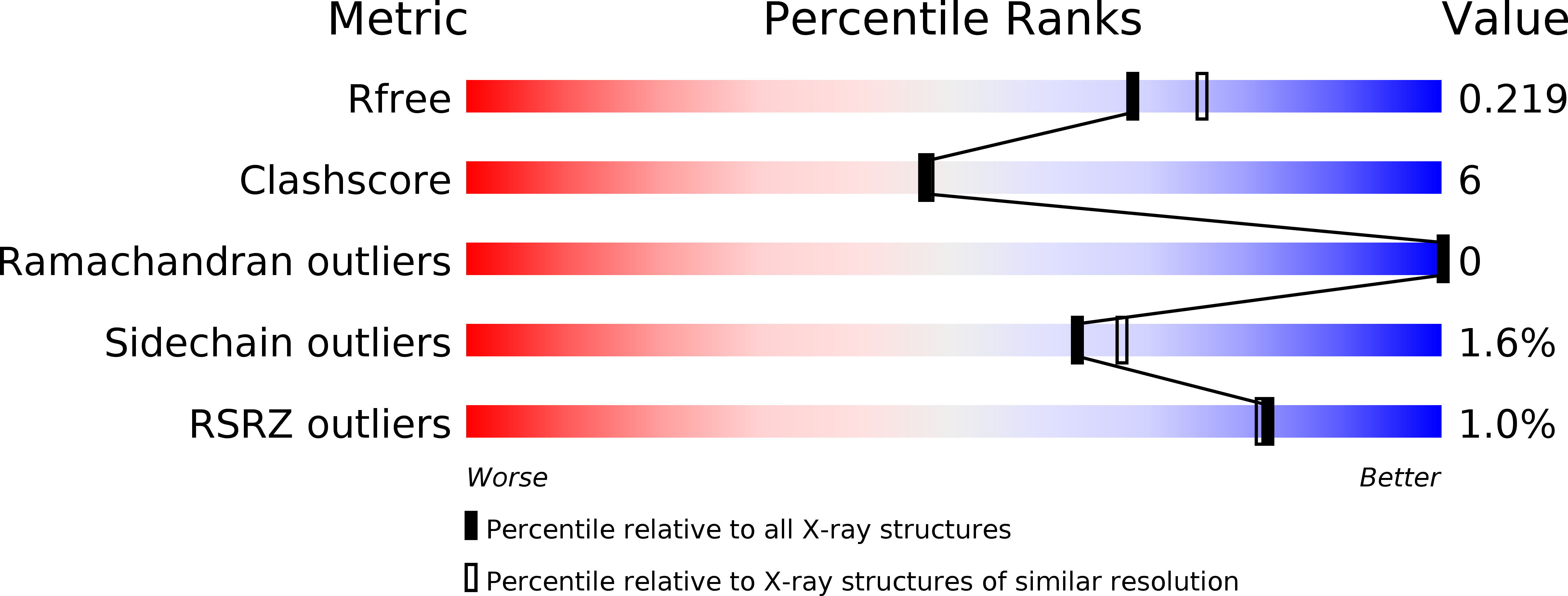
5GZA - PubMed Abstract:
The 'pseudokinase' SgK196 is a protein O-mannose kinase (POMK) that catalyzes an essential phosphorylation step during biosynthesis of the laminin-binding glycan on α-dystroglycan. However, the catalytic mechanism underlying this activity remains elusive. Here we present the crystal structure of Danio rerio POMK in complex with Mg 2+ ions, ADP, aluminum fluoride, and the GalNAc-β3-GlcNAc-β4-Man trisaccharide substrate, thereby providing a snapshot of the catalytic transition state of this unusual kinase. The active site of POMK is established by residues located in non-canonical positions and is stabilized by a disulfide bridge. GalNAc-β3-GlcNAc-β4-Man is recognized by a surface groove, and the GalNAc-β3-GlcNAc moiety mediates the majority of interactions with POMK. Expression of various POMK mutants in POMK knockout cells further validated the functional requirements of critical residues. Our results provide important insights into the ability of POMK to function specifically as a glycan kinase, and highlight the structural diversity of the human kinome.
Organizational Affiliation:
The State Key Laboratory of Protein and Plant Gene Research, School of Life Sciences, Peking University, Beijing, China.