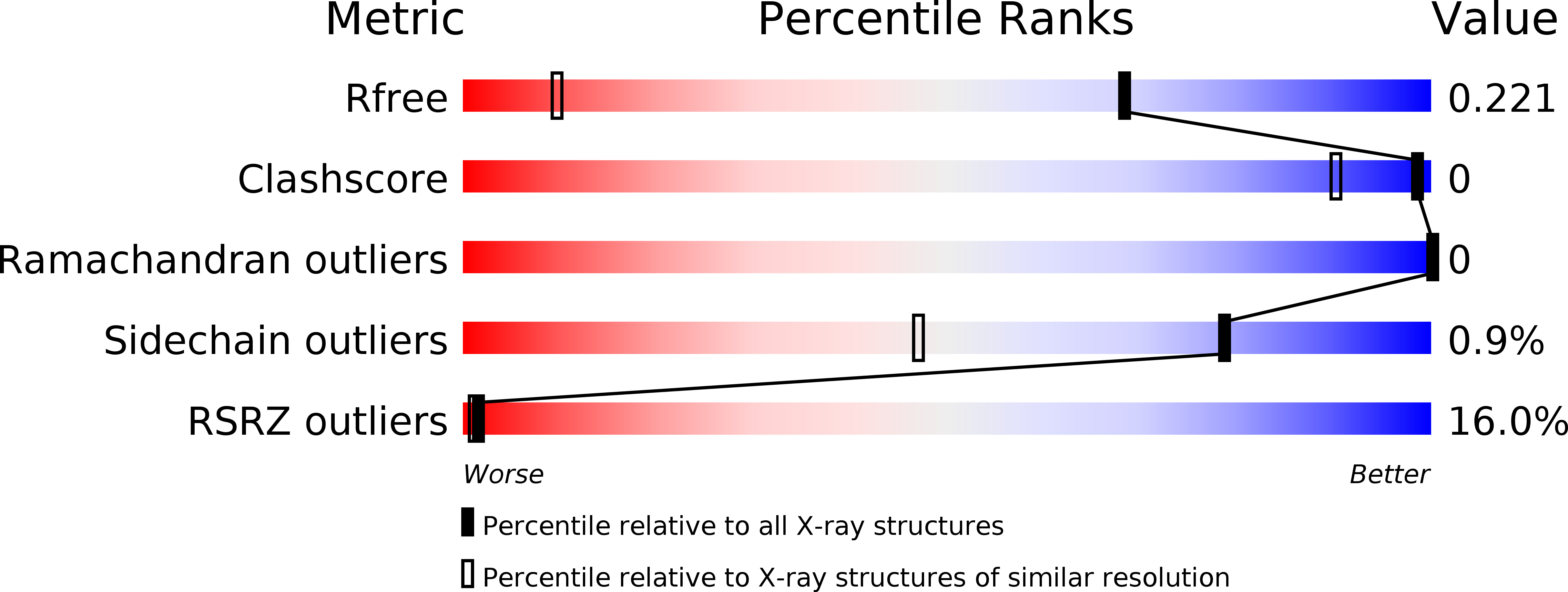
PanDDA analysis group deposition
Talon, R., Krojer, T., Fairhead, M., Sethi, R., Bradley, A.R., Aimon, A., Collins, P., Brandao-Neto, J., Douangamath, A., Wright, N., MacLean, E., Renjie, Z., Dias, A., Brennan, P.E., Bountra, C., Arrowsmith, C.H., Edwards, A., von Delft, F.To be published.