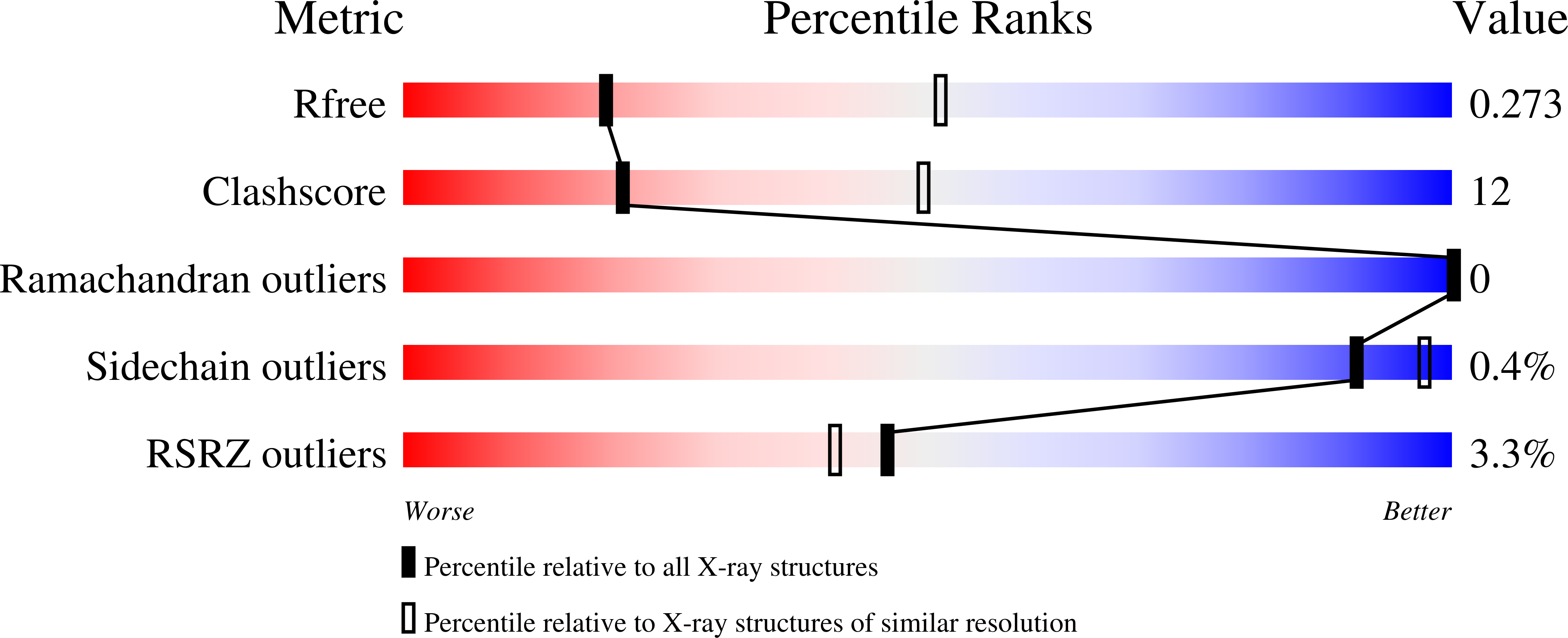
KAT2A coupled with the alpha-KGDH complex acts as a histone H3 succinyltransferase.
Wang, Y., Guo, Y.R., Liu, K., Yin, Z., Liu, R., Xia, Y., Tan, L., Yang, P., Lee, J.H., Li, X.J., Hawke, D., Zheng, Y., Qian, X., Lyu, J., He, J., Xing, D., Tao, Y.J., Lu, Z.(2017) Nature 552: 273-277
- PubMed: 29211711
- DOI: https://doi.org/10.1038/nature25003
- Primary Citation of Related Structures:
5TRL, 5TRM - PubMed Abstract:
Histone modifications, such as the frequently occurring lysine succinylation, are central to the regulation of chromatin-based processes. However, the mechanism and functional consequences of histone succinylation are unknown. Here we show that the α-ketoglutarate dehydrogenase (α-KGDH) complex is localized in the nucleus in human cell lines and binds to lysine acetyltransferase 2A (KAT2A, also known as GCN5) in the promoter regions of genes. We show that succinyl-coenzyme A (succinyl-CoA) binds to KAT2A. The crystal structure of the catalytic domain of KAT2A in complex with succinyl-CoA at 2.3 Å resolution shows that succinyl-CoA binds to a deep cleft of KAT2A with the succinyl moiety pointing towards the end of a flexible loop 3, which adopts different structural conformations in succinyl-CoA-bound and acetyl-CoA-bound forms. Site-directed mutagenesis indicates that tyrosine 645 in this loop has an important role in the selective binding of succinyl-CoA over acetyl-CoA. KAT2A acts as a succinyltransferase and succinylates histone H3 on lysine 79, with a maximum frequency around the transcription start sites of genes. Preventing the α-KGDH complex from entering the nucleus, or expression of KAT2A(Tyr645Ala), reduces gene expression and inhibits tumour cell proliferation and tumour growth. These findings reveal an important mechanism of histone modification and demonstrate that local generation of succinyl-CoA by the nuclear α-KGDH complex coupled with the succinyltransferase activity of KAT2A is instrumental in histone succinylation, tumour cell proliferation, and tumour development.
Organizational Affiliation:
Brain Tumor Center, Department of Neuro-Oncology, The University of Texas MD Anderson Cancer Center, Houston, Texas 77030, USA.