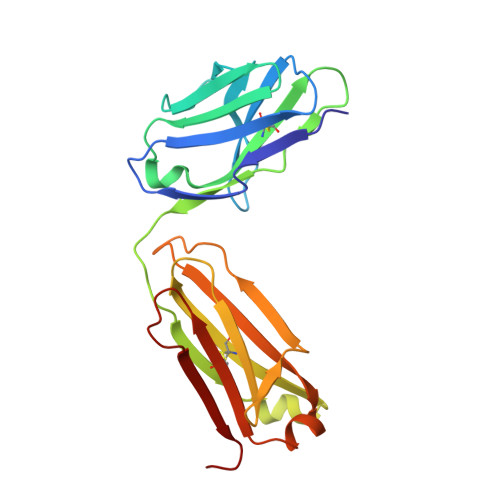
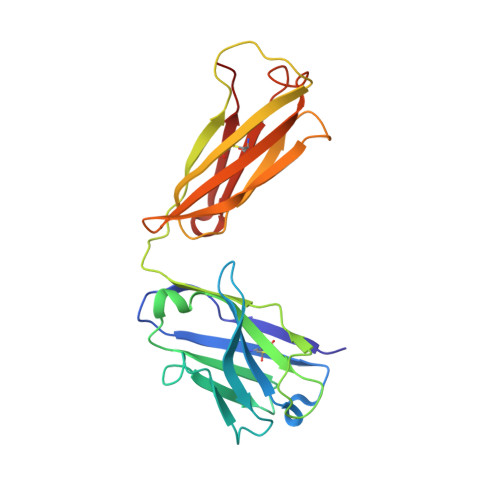
Crystal structure of the human angiotensin II type 2 receptor bound to an angiotensin II analog.
Asada, H., Horita, S., Hirata, K., Shiroishi, M., Shiimura, Y., Iwanari, H., Hamakubo, T., Shimamura, T., Nomura, N., Kusano-Arai, O., Uemura, T., Suno, C., Kobayashi, T., Iwata, S.(2018) Nat Struct Mol Biol 25: 570-576
- PubMed: 29967536
- DOI: https://doi.org/10.1038/s41594-018-0079-8
- Primary Citation of Related Structures:
5XJM, 5XLI - PubMed Abstract:
Angiotensin II (AngII) plays a central role in regulating human blood pressure, which is mainly mediated by interactions between AngII and the G-protein-coupled receptors (GPCRs) AngII type 1 receptor (AT 1 R) and AngII type 2 receptor (AT 2 R). We have solved the crystal structure of human AT 2 R binding the peptide ligand [Sar 1 , Ile 8 ]AngII and its specific antibody at 3.2-Å resolution. [Sar 1 , Ile 8 ]AngII interacts with both the 'core' binding domain, where the small-molecule ligands of AT 1 R and AT 2 R bind, and the 'extended' binding domain, which is equivalent to the allosteric modulator binding site of muscarinic acetylcholine receptor. We generated an antibody fragment to stabilize the extended binding domain that functions as a positive allosteric modulator. We also identified a signature positively charged cluster, which is conserved among peptide-binding receptors, to locate C termini at the bottom of the binding pocket. The reported results should help with designing ligands for angiotensin receptors and possibly to other peptide GPCRs.
Organizational Affiliation:
Department of Cell Biology, Graduate School of Medicine, Kyoto University, Kyoto, Japan.