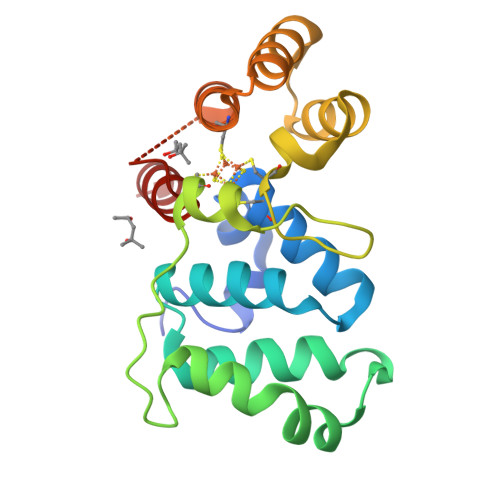
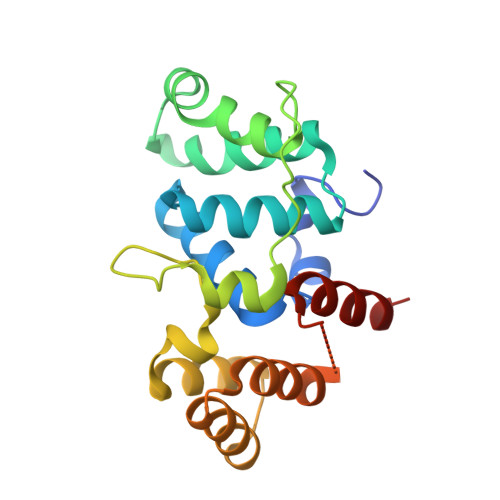
Yeast require redox switching in DNA primase.
O'Brien, E., Salay, L.E., Epum, E.A., Friedman, K.L., Chazin, W.J., Barton, J.K.(2018) Proc Natl Acad Sci U S A 115: 13186-13191
- PubMed: 30541886
- DOI: https://doi.org/10.1073/pnas.1810715115
- Primary Citation of Related Structures:
6DI2, 6DI6, 6DTV, 6DTZ, 6DU0 - PubMed Abstract:
Eukaryotic DNA primases contain a [4Fe4S] cluster in the C-terminal domain of the p58 subunit (p58C) that affects substrate affinity but is not required for catalysis. We show that, in yeast primase, the cluster serves as a DNA-mediated redox switch governing DNA binding, just as in human primase. Despite a different structural arrangement of tyrosines to facilitate electron transfer between the DNA substrate and [4Fe4S] cluster, in yeast, mutation of tyrosines Y395 and Y397 alters the same electron transfer chemistry and redox switch. Mutation of conserved tyrosine 395 diminishes the extent of p58C participation in normal redox-switching reactions, whereas mutation of conserved tyrosine 397 causes oxidative cluster degradation to the [3Fe4S] + species during p58C redox signaling. Switching between oxidized and reduced states in the presence of the Y397 mutations thus puts primase [4Fe4S] cluster integrity and function at risk. Consistent with these observations, we find that yeast tolerate mutations to Y395 in p58C, but the single-residue mutation Y397L in p58C is lethal. Our data thus show that a constellation of tyrosines for protein-DNA electron transfer mediates the redox switch in eukaryotic primases and is required for primase function in vivo.
Organizational Affiliation:
Division of Chemistry and Chemical Engineering, California Institute of Technology, Pasadena, CA 91125.