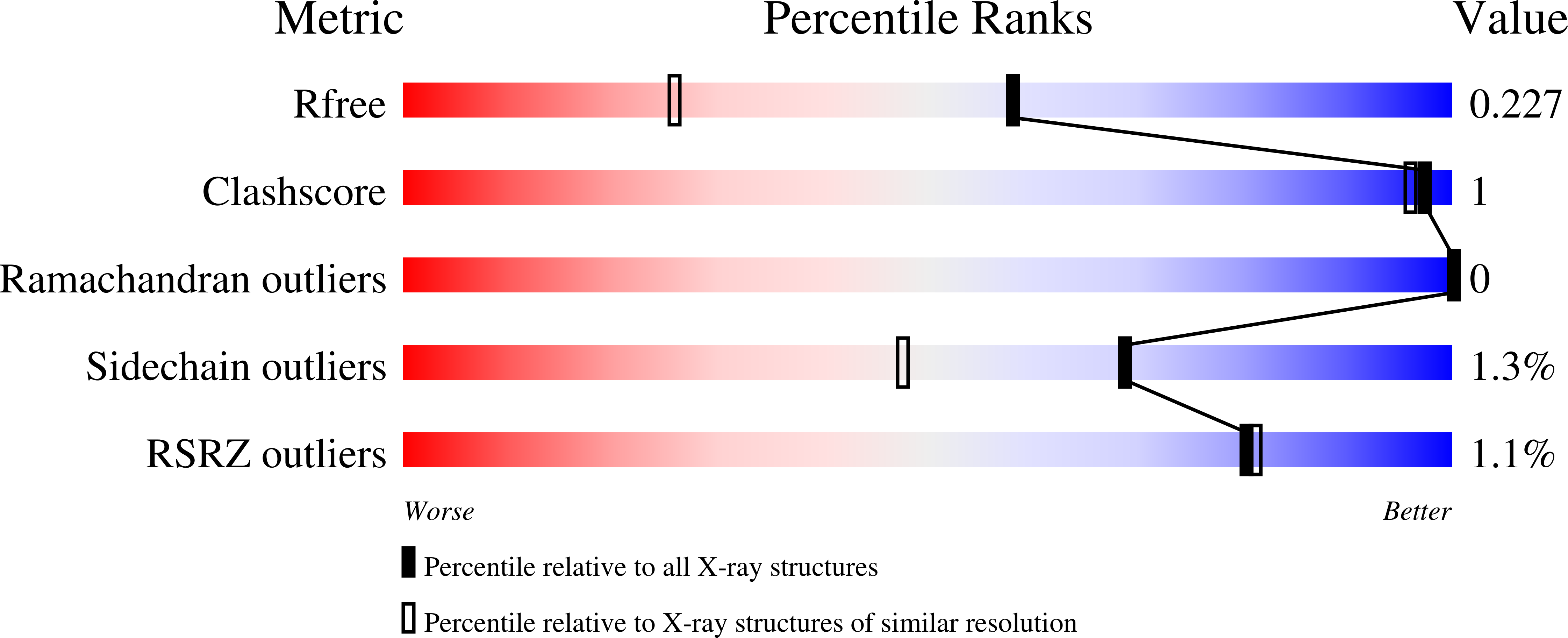
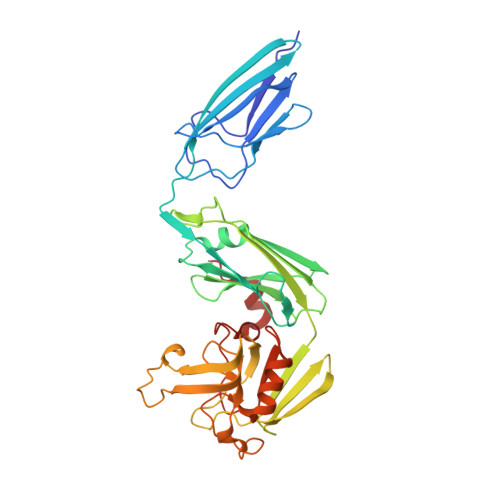
Targeting the Mycobacterium tuberculosis transpeptidase LdtMt2with cysteine-reactive inhibitors including ebselen.
de Munnik, M., Lohans, C.T., Lang, P.A., Langley, G.W., Malla, T.R., Tumber, A., Schofield, C.J., Brem, J.(2019) Chem Commun (Camb) 55: 10214-10217
- PubMed: 31380528
- DOI: https://doi.org/10.1039/c9cc04145a
- Primary Citation of Related Structures:
6RLG, 6RRM - PubMed Abstract:
The l,d-transpeptidases (Ldts) are promising antibiotic targets for treating tuberculosis. We report screening of cysteine-reactive inhibitors against LdtMt2 from Mycobacterium tuberculosis. Structural studies on LdtMt2 with potent inhibitor ebselen reveal opening of the benzisoselenazolone ring by a nucleophilic cysteine, forming a complex involving extensive hydrophobic interactions with a substrate-binding loop.
Organizational Affiliation:
Chemistry Research Laboratory, Department of Chemistry, University of Oxford, Oxford, OX1 3TA, UK. christopher.schofield@chem.ox.ac.uk jurgen.brem@chem.ox.ac.uk.