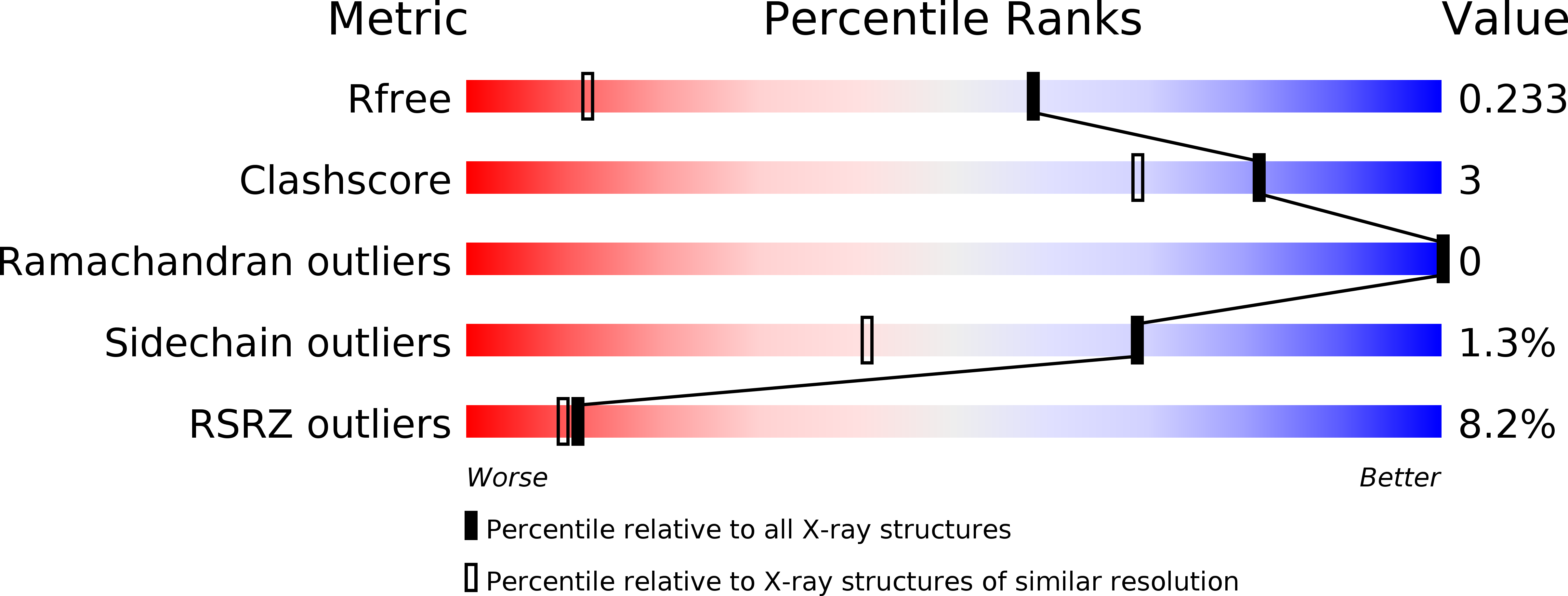
Crystal Structure of 1-methylindoline-2,3-dione covalently bound to the PH domain of Bruton's tyrosine kinase mutant R28C
Brear, P., Fischer, G., May, M., Pantelejevs, T., Mathieu, R., Rossmann, M., Wagstaff, J., Blaszczyk, B., Hyvonen, M.To be published.