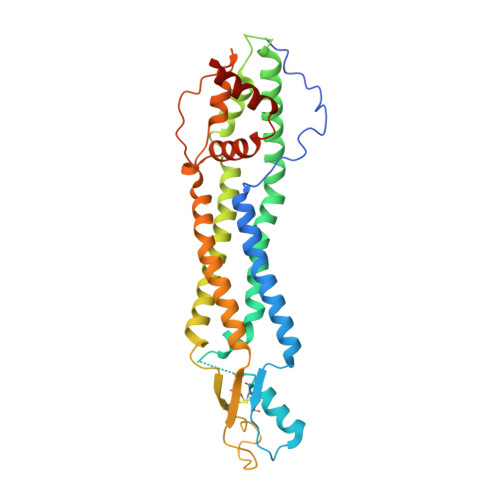
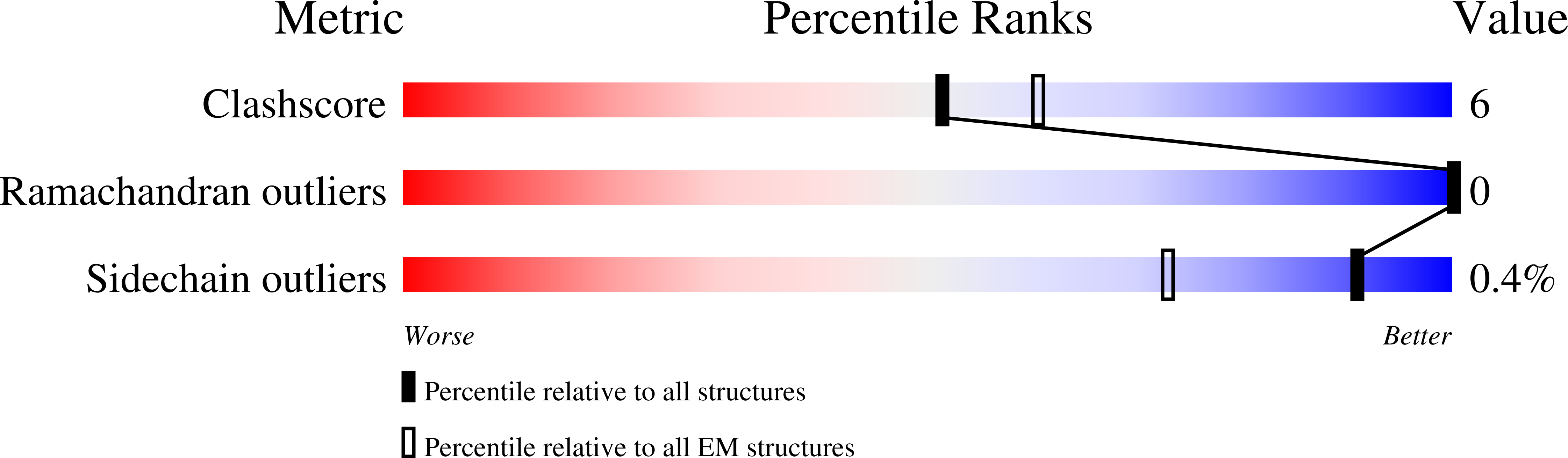The Cryo-EM structure of pannexin 1 reveals unique motifs for ion selection and inhibition.
Michalski, K., Syrjanen, J.L., Henze, E., Kumpf, J., Furukawa, H., Kawate, T.(2020) Elife 9
- PubMed: 32048993
- DOI: https://doi.org/10.7554/eLife.54670
- Primary Citation of Related Structures:
6VD7 - PubMed Abstract:
Pannexins are large-pore forming channels responsible for ATP release under a variety of physiological and pathological conditions. Although predicted to share similar membrane topology with other large-pore forming proteins such as connexins, innexins, and LRRC8, pannexins have minimal sequence similarity to these protein families. Here, we present the cryo-EM structure of a frog pannexin 1 (Panx1) channel at 3.0 Å. We find that Panx1 protomers harbor four transmembrane helices similar in arrangement to other large-pore forming proteins but assemble as a heptameric channel with a unique constriction formed by Trp74 in the first extracellular loop. Mutating Trp74 or the nearby Arg75 disrupt ion selectivity, whereas altering residues in the hydrophobic groove formed by the two extracellular loops abrogates channel inhibition by carbenoxolone. Our structural and functional study establishes the extracellular loops as important structural motifs for ion selectivity and channel inhibition in Panx1.
Organizational Affiliation:
Department of Molecular Medicine, Cornell University, Ithaca, United States.