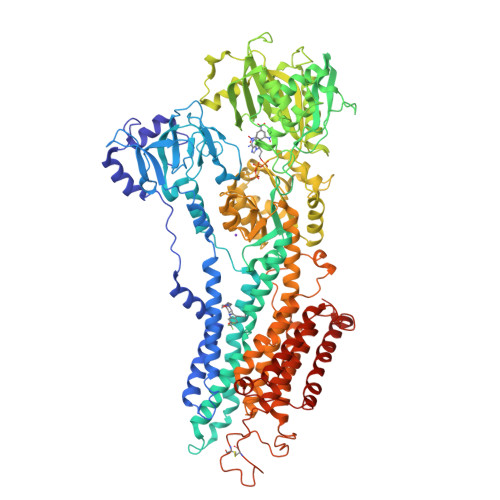
Blockade of Oncogenic NOTCH1 with the SERCA Inhibitor CAD204520 in T Cell Acute Lymphoblastic Leukemia.
Marchesini, M., Gherli, A., Montanaro, A., Patrizi, L., Sorrentino, C., Pagliaro, L., Rompietti, C., Kitara, S., Heit, S., Olesen, C.E., Moller, J.V., Savi, M., Bocchi, L., Vilella, R., Rizzi, F., Baglione, M., Rastelli, G., Loiacono, C., La Starza, R., Mecucci, C., Stegmaier, K., Aversa, F., Stilli, D., Lund Winther, A.M., Sportoletti, P., Bublitz, M., Dalby-Brown, W., Roti, G.(2020) Cell Chem Biol 27: 678
- PubMed: 32386594
- DOI: https://doi.org/10.1016/j.chembiol.2020.04.002
- Primary Citation of Related Structures:
6YAA - PubMed Abstract:
The identification of SERCA (sarco/endoplasmic reticulum calcium ATPase) as a target for modulating gain-of-function NOTCH1 mutations in Notch-dependent cancers has spurred the development of this compound class for cancer therapeutics. Despite the innate toxicity challenge associated with SERCA inhibition, we identified CAD204520, a small molecule with better drug-like properties and reduced off-target Ca 2+ toxicity compared with the SERCA inhibitor thapsigargin. In this work, we describe the properties and complex structure of CAD204520 and show that CAD204520 preferentially targets mutated over wild-type NOTCH1 proteins in T cell acute lymphoblastic leukemia (T-ALL) and mantle cell lymphoma (MCL). Uniquely among SERCA inhibitors, CAD204520 suppresses NOTCH1-mutated leukemic cells in a T-ALL xenografted model without causing cardiac toxicity. This study supports the development of SERCA inhibitors for Notch-dependent cancers and extends their application to cases with isolated mutations in the PEST degradation domain of NOTCH1, such as MCL or chronic lymphocytic leukemia (CLL).
Organizational Affiliation:
University of Parma, Department of Medicine and Surgery, Parma 43126, Italy.