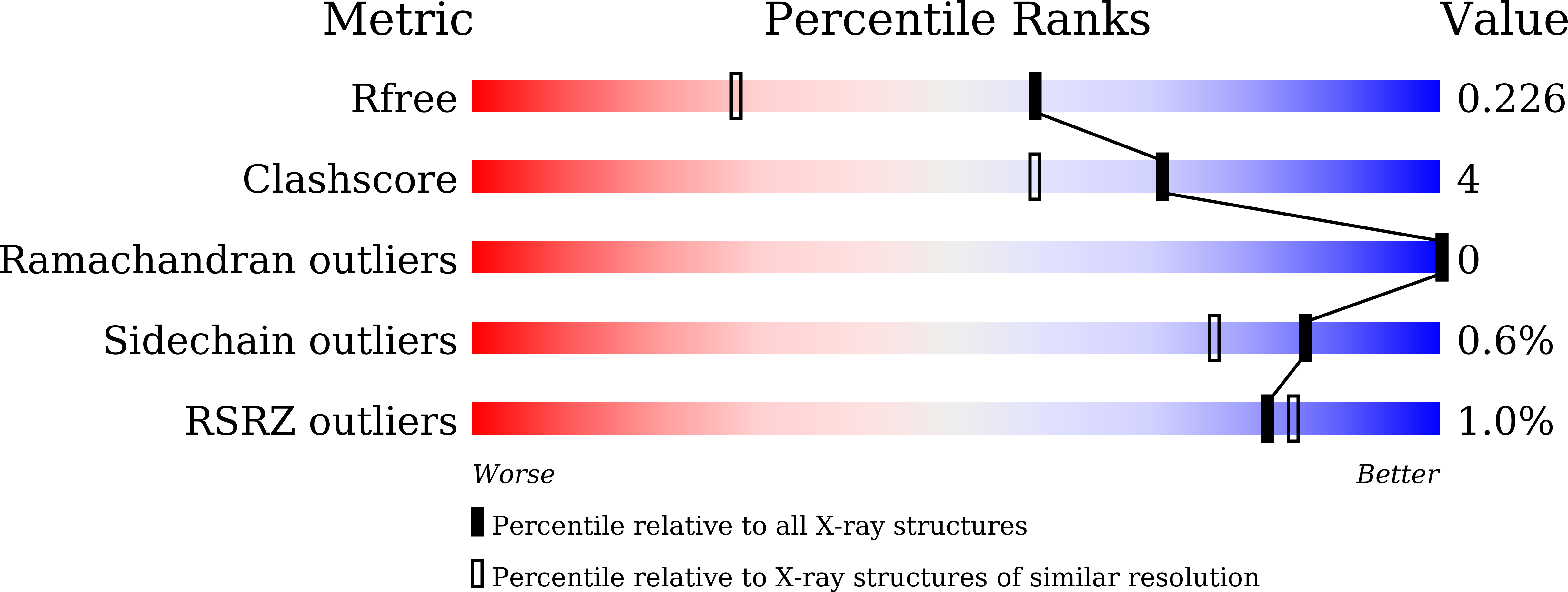
Molecular basis for reduced cleavage activity and drug resistance in D30N HIV-1 protease.
Bihani, S.C., Gupta, G.D., Hosur, M.V.(2021) J Biomol Struct Dyn : 1-9
- PubMed: 34609269
- DOI: https://doi.org/10.1080/07391102.2021.1982007
- Primary Citation of Related Structures:
7DOZ, 7DPQ - PubMed Abstract:
Nelfinavir is one of the FDA-approved HIV-1 protease inhibitors and a part of highly active anti-retroviral therapy (HAART) for the treatment of HIV-AIDS. Nelfinavir was the first HIV-1 protease inhibitor to be approved as a paediatric formulation. The application of HAART had resulted in significant improvement in the lives of AIDS patients. However, the emergence of drug resistance in HIV-1 protease has limited the use of many of these drugs including nelfinavir. A unique mutation observed frequently in patients treated with nelfinavir is D30N as it is selected exclusively by nelfinavir. The D30N mutation imparts very high resistance to nelfinavir but unlike other primary mutations does not give cross-resistance to the majority of other drugs. D30N mutation also significantly reduces cleavage activity of HIV-1 protease and affects viral fitness. Here, we have determined crystal structures of D30N HIV-1 protease in unliganded form and in complex with nelfinavir. These structures provide the rationale for reduced cleavage activity and the molecular basis of drug resistance induced by D30N mutation. The loss of coulombic interaction part of a crucial hydrogen bond between the drug and the protease is likely to play a major role in reduced affinity and resistance towards nelfinavir. The decreased catalytic activity of D30N HIV-1 protease due to altered interaction with the substrates and reduced stability of folding core may be the reason for the reduced replicative capacity of the virus harboring mutant HIV-1 protease.Communicated by Ramaswamy H. Sarma.
Organizational Affiliation:
Protein Crystallography Section, Radiation Biology & Health Sciences Division, Bhabha Atomic Research Centre, Mumbai,India.