A natural mutation between SARS-CoV-2 and SARS-CoV determines neutralization by a cross-reactive antibody.
Wu, N.C., Yuan, M., Bangaru, S., Huang, D., Zhu, X., Lee, C.D., Turner, H.L., Peng, L., Yang, L., Burton, D.R., Nemazee, D., Ward, A.B., Wilson, I.A.(2020) PLoS Pathog 16: e1009089-e1009089
- PubMed: 33275640
- DOI: https://doi.org/10.1371/journal.ppat.1009089
- Primary Citation of Related Structures:
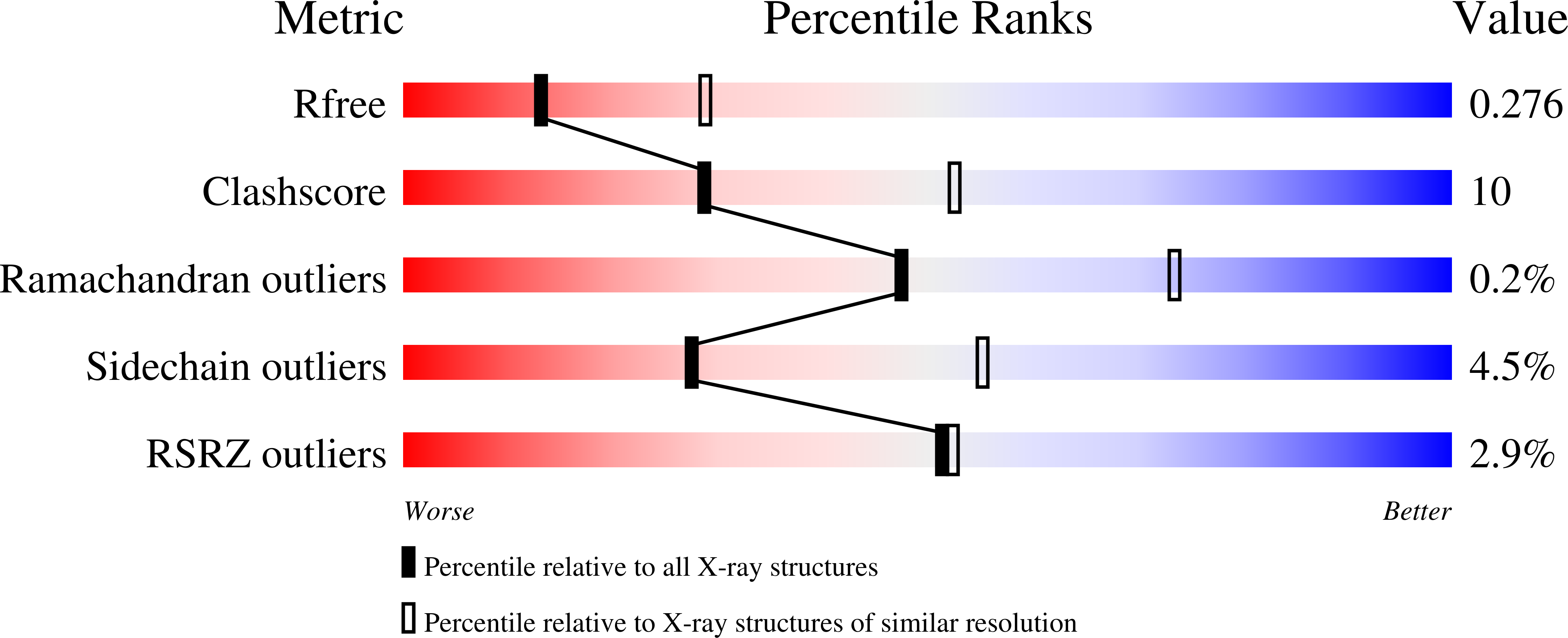
7JN5 - PubMed Abstract:
Epitopes that are conserved among SARS-like coronaviruses are attractive targets for design of cross-reactive vaccines and therapeutics. CR3022 is a SARS-CoV neutralizing antibody to a highly conserved epitope on the receptor binding domain (RBD) on the spike protein that is able to cross-react with SARS-CoV-2, but with lower affinity. Using x-ray crystallography, mutagenesis, and binding experiments, we illustrate that of four amino acid differences in the CR3022 epitope between SARS-CoV-2 and SARS-CoV, a single mutation P384A fully determines the affinity difference. CR3022 does not neutralize SARS-CoV-2, but the increased affinity to SARS-CoV-2 P384A mutant now enables neutralization with a similar potency to SARS-CoV. We further investigated CR3022 interaction with the SARS-CoV spike protein by negative-stain EM and cryo-EM. Three CR3022 Fabs bind per trimer with the RBD observed in different up-conformations due to considerable flexibility of the RBD. In one of these conformations, quaternary interactions are made by CR3022 to the N-terminal domain (NTD) of an adjacent subunit. Overall, this study provides insights into antigenic variation and potential cross-neutralizing epitopes on SARS-like viruses.
Organizational Affiliation:
Department of Biochemistry, University of Illinois at Urbana-Champaign, Urbana, IL, United States of America.