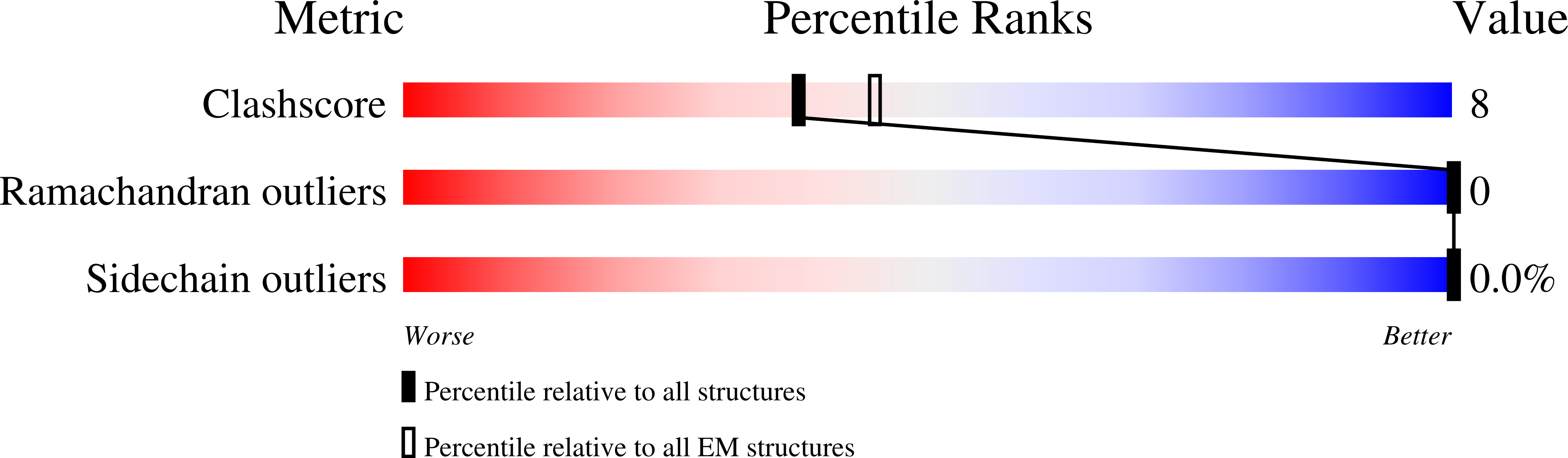Structures of synthetic nanobody-SARS-CoV-2 receptor-binding domain complexes reveal distinct sites of interaction.
Ahmad, J., Jiang, J., Boyd, L.F., Zeher, A., Huang, R., Xia, D., Natarajan, K., Margulies, D.H.(2021) J Biol Chem 297: 101202-101202
- PubMed: 34537245
- DOI: https://doi.org/10.1016/j.jbc.2021.101202
- Primary Citation of Related Structures:
7KGJ, 7KGK, 7KLW, 7MFU, 7MFV, 7N0G, 7N0H - PubMed Abstract:
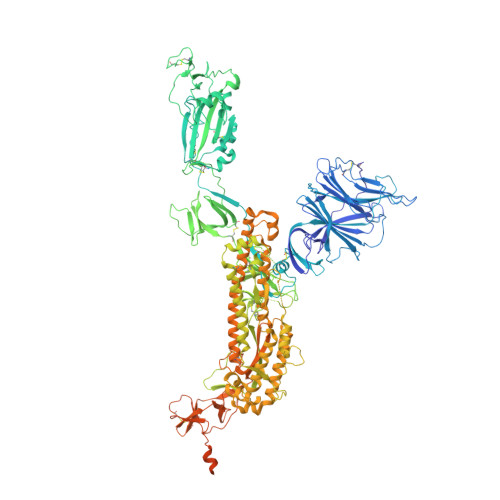
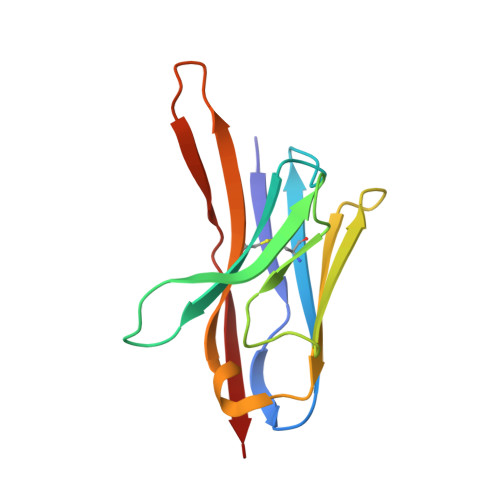
Combating the worldwide spread of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and the emergence of new variants demands understanding of the structural basis of the interaction of antibodies with the SARS-CoV-2 receptor-binding domain (RBD). Here, we report five X-ray crystal structures of sybodies (synthetic nanobodies) including those of binary and ternary complexes of Sb16-RBD, Sb45-RBD, Sb14-RBD-Sb68, and Sb45-RBD-Sb68, as well as unliganded Sb16. These structures reveal that Sb14, Sb16, and Sb45 bind the RBD at the angiotensin-converting enzyme 2 interface and that the Sb16 interaction is accompanied by a large conformational adjustment of complementarity-determining region 2. In contrast, Sb68 interacts at the periphery of the SARS-CoV-2 RBD-angiotensin-converting enzyme 2 interface. We also determined cryo-EM structures of Sb45 bound to the SARS-CoV-2 spike protein. Superposition of the X-ray structures of sybodies onto the trimeric spike protein cryo-EM map indicates that some sybodies may bind in both "up" and "down" configurations, but others may not. Differences in sybody recognition of several recently identified RBD variants are explained by these structures.
Organizational Affiliation:
Molecular Biology Section, Laboratory of Immune System Biology, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Maryland, USA.