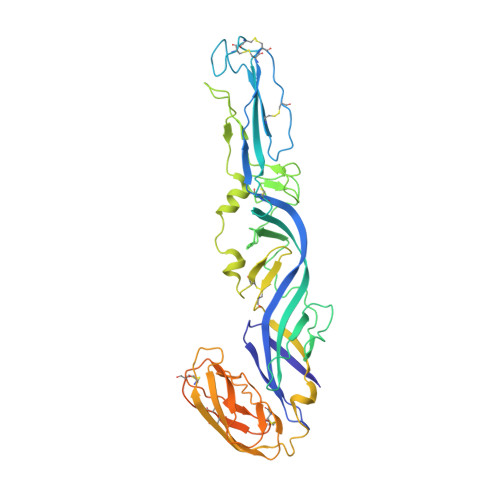
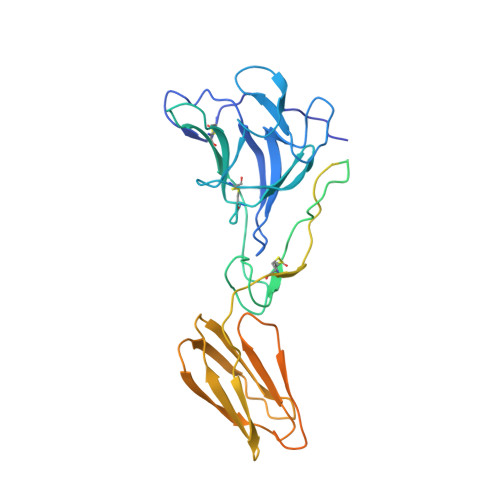
Cryo-EM structures of alphavirus conformational intermediates in low pH-triggered prefusion states.
Chen, C.L., Klose, T., Sun, C., Kim, A.S., Buda, G., Rossmann, M.G., Diamond, M.S., Klimstra, W.B., Kuhn, R.J.(2022) Proc Natl Acad Sci U S A 119: e2114119119-e2114119119
- PubMed: 35867819
- DOI: https://doi.org/10.1073/pnas.2114119119
- Primary Citation of Related Structures:
7N69, 7N6A - PubMed Abstract:
Alphaviruses can cause severe human arthritis and encephalitis. During virus infection, structural changes of viral glycoproteins in the acidified endosome trigger virus-host membrane fusion for delivery of the capsid core and RNA genome into the cytosol to initiate virus translation and replication. However, mechanisms by which E1 and E2 glycoproteins rearrange in this process remain unknown. Here, we investigate prefusion cryoelectron microscopy (cryo-EM) structures of eastern equine encephalitis virus (EEEV) under acidic conditions. With models fitted into the low-pH cryo-EM maps, we suggest that E2 dissociates from E1, accompanied by a rotation (∼60°) of the E2-B domain (E2-B) to expose E1 fusion loops. Cryo-EM reconstructions of EEEV bound to a protective antibody at acidic and neutral pH suggest that stabilization of E2-B prevents dissociation of E2 from E1. These findings reveal conformational changes of the glycoprotein spikes in the acidified host endosome. Stabilization of E2-B may provide a strategy for antiviral agent development.
Organizational Affiliation:
Department of Biological Sciences, Purdue University, West Lafayette, IN 47907.