X-Ray structure of Rhodobacter sphaeroides reaction center with M197 Phe-His substitution clarifies properties of the mutant complex
Gabdulkhakov, A.G., Selikhanov, G.K., Guenther, S., Meents, A., Fufina, T.Y., Vasilieva, L.G.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Reaction center protein H chain | A [auth H] | 241 | Cereibacter sphaeroides | Mutation(s): 0 Gene Names: puhA Membrane Entity: Yes | 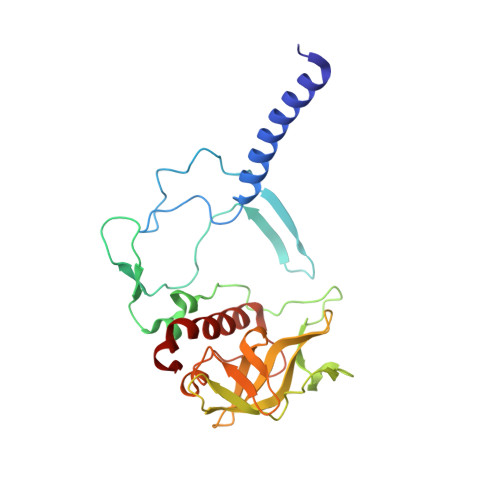 |
UniProt | |||||
Find proteins for P0C0Y7 (Cereibacter sphaeroides) Explore P0C0Y7 Go to UniProtKB: P0C0Y7 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0C0Y7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Reaction center protein L chain | B [auth L] | 281 | Cereibacter sphaeroides | Mutation(s): 1 Gene Names: pufL Membrane Entity: Yes | 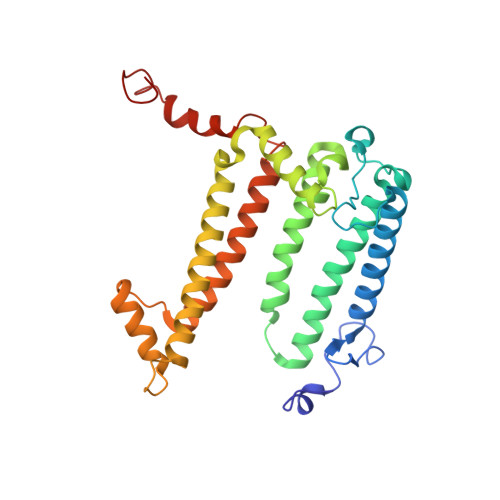 |
UniProt | |||||
Find proteins for P0C0Y8 (Cereibacter sphaeroides) Explore P0C0Y8 Go to UniProtKB: P0C0Y8 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0C0Y8 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Reaction center protein M chain | C [auth M] | 303 | Cereibacter sphaeroides | Mutation(s): 2 Gene Names: pufM Membrane Entity: Yes | 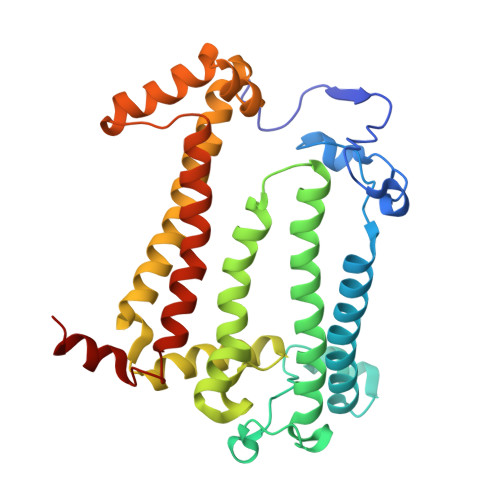 |
UniProt | |||||
Find proteins for P0C0Y9 (Cereibacter sphaeroides) Explore P0C0Y9 Go to UniProtKB: P0C0Y9 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0C0Y9 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 10 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| BCL (Subject of Investigation/LOI) Query on BCL | I [auth L], J [auth L], S [auth M], T [auth M] | BACTERIOCHLOROPHYLL A C55 H74 Mg N4 O6 DSJXIQQMORJERS-AGGZHOMASA-M |  | ||
| BPH (Subject of Investigation/LOI) Query on BPH | K [auth L], U [auth M] | BACTERIOPHEOPHYTIN A C55 H76 N4 O6 KWOZSBGNAHVCKG-SZQBJALDSA-N |  | ||
| U10 Query on U10 | Q [auth M] | UBIQUINONE-10 C59 H90 O4 ACTIUHUUMQJHFO-UPTCCGCDSA-N |  | ||
| SPN Query on SPN | W [auth M] | SPEROIDENONE C41 H70 O2 GWQAMGYOEYXWJF-YCDPMLDASA-N |  | ||
| NKP Query on NKP | M, O [auth M] | (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E)-octadec-9-enoate C21 H41 O7 P WRGQSWVCFNIUNZ-SQUSKLHYSA-N |  | ||
| OLC Query on OLC | G [auth L] | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate C21 H40 O4 RZRNAYUHWVFMIP-GDCKJWNLSA-N |  | ||
| LDA Query on LDA | D [auth H], F [auth H], X [auth M], Y [auth M] | LAURYL DIMETHYLAMINE-N-OXIDE C14 H31 N O SYELZBGXAIXKHU-UHFFFAOYSA-N |  | ||
| EDO Query on EDO | AA [auth M] | 1,2-ETHANEDIOL C2 H6 O2 LYCAIKOWRPUZTN-UHFFFAOYSA-N |  | ||
| FE Query on FE | V [auth M] | FE (III) ION Fe VTLYFUHAOXGGBS-UHFFFAOYSA-N |  | ||
| UNL Query on UNL | E [auth H] H [auth L] L N [auth M] P [auth M] | Unknown ligand VTLYFUHAOXGGBS-UHFFFAOYSA-N | |||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 99.97 | α = 90 |
| b = 99.97 | β = 90 |
| c = 239.22 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| PHENIX | refinement |
| XDS | data reduction |
| XSCALE | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Russian Foundation for Basic Research | Russian Federation | 18-02-40008_mega |