Biocatalytically active and stable cross-linked enzyme crystals of halohydrin dehalogenase HheG by protein engineering
Staar, M., Henke, S., Blankenfeldt, W., Schallmey, A.(2022) ChemCatChem
Experimental Data Snapshot
Starting Model: experimental
View more details
(2022) ChemCatChem
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Putative oxidoreductase | 282 | Ilumatobacter coccineus YM16-304 | Mutation(s): 1 Gene Names: YM304_35350 | 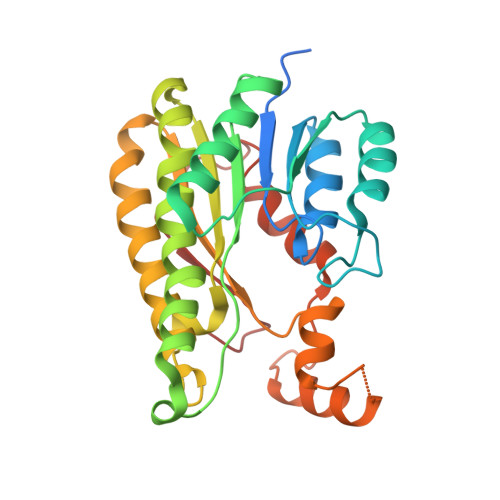 | |
UniProt | |||||
Find proteins for A0A6C7EF96 (Ilumatobacter coccineus (strain NBRC 103263 / KCTC 29153 / YM16-304)) Explore A0A6C7EF96 Go to UniProtKB: A0A6C7EF96 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A6C7EF96 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| ME7 (Subject of Investigation/LOI) Query on ME7 | EA [auth F], K [auth A], LA [auth H], OA [auth I], Q [auth B] | 1,1'-ethane-1,2-diylbis(1H-pyrrole-2,5-dione) C10 H8 N2 O4 PUKLCKVOVCZYKF-UHFFFAOYSA-N |  | ||
| SO4 Query on SO4 | AA [auth E] BA [auth E] CA [auth E] DA [auth E] FA [auth F] | SULFATE ION O4 S QAOWNCQODCNURD-UHFFFAOYSA-L |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 195.08 | α = 90 |
| b = 195.08 | β = 90 |
| c = 195.064 | γ = 120 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| XDS | data reduction |
| Aimless | data scaling |
| PDB_EXTRACT | data extraction |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| German Research Foundation (DFG) | Germany | 281361126/GRK2223 |
| German Research Foundation (DFG) | Germany | SPP 1934, SCHA 1745/2-2 |