Structural basis of regulated m 7 G tRNA modification by METTL1-WDR4.
Li, J., Wang, L., Hahn, Q., Nowak, R.P., Viennet, T., Orellana, E.A., Roy Burman, S.S., Yue, H., Hunkeler, M., Fontana, P., Wu, H., Arthanari, H., Fischer, E.S., Gregory, R.I.(2023) Nature 613: 391-397
- PubMed: 36599985
- DOI: https://doi.org/10.1038/s41586-022-05566-4
- Primary Citation of Related Structures:
7U20, 8CTH, 8CTI - PubMed Abstract:
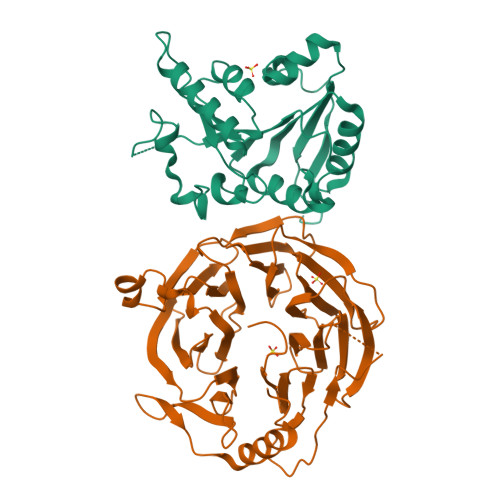
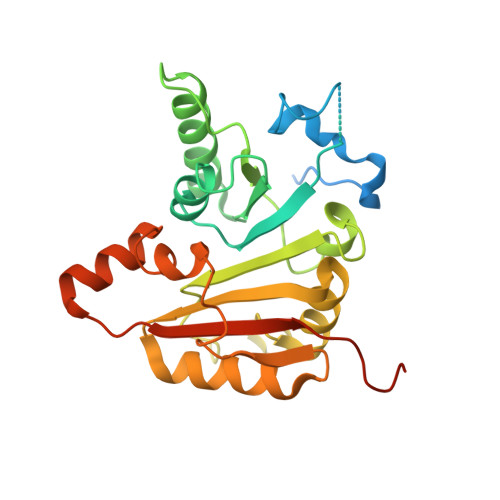
Chemical modifications of RNA have key roles in many biological processes 1-3 . N 7 -methylguanosine (m 7 G) is required for integrity and stability of a large subset of tRNAs 4-7 . The methyltransferase 1-WD repeat-containing protein 4 (METTL1-WDR4) complex is the methyltransferase that modifies G46 in the variable loop of certain tRNAs, and its dysregulation drives tumorigenesis in numerous cancer types 8-14 . Mutations in WDR4 cause human developmental phenotypes including microcephaly 15-17 . How METTL1-WDR4 modifies tRNA substrates and is regulated remains elusive 18 . Here we show, through structural, biochemical and cellular studies of human METTL1-WDR4, that WDR4 serves as a scaffold for METTL1 and the tRNA T-arm. Upon tRNA binding, the αC region of METTL1 transforms into a helix, which together with the α6 helix secures both ends of the tRNA variable loop. Unexpectedly, we find that the predicted disordered N-terminal region of METTL1 is part of the catalytic pocket and essential for methyltransferase activity. Furthermore, we reveal that S27 phosphorylation in the METTL1 N-terminal region inhibits methyltransferase activity by locally disrupting the catalytic centre. Our results provide a molecular understanding of tRNA substrate recognition and phosphorylation-mediated regulation of METTL1-WDR4, and reveal the presumed disordered N-terminal region of METTL1 as a nexus of methyltransferase activity.
Organizational Affiliation:
Stem Cell Program, Division of Hematology/Oncology, Boston Children's Hospital, Boston, MA, USA.