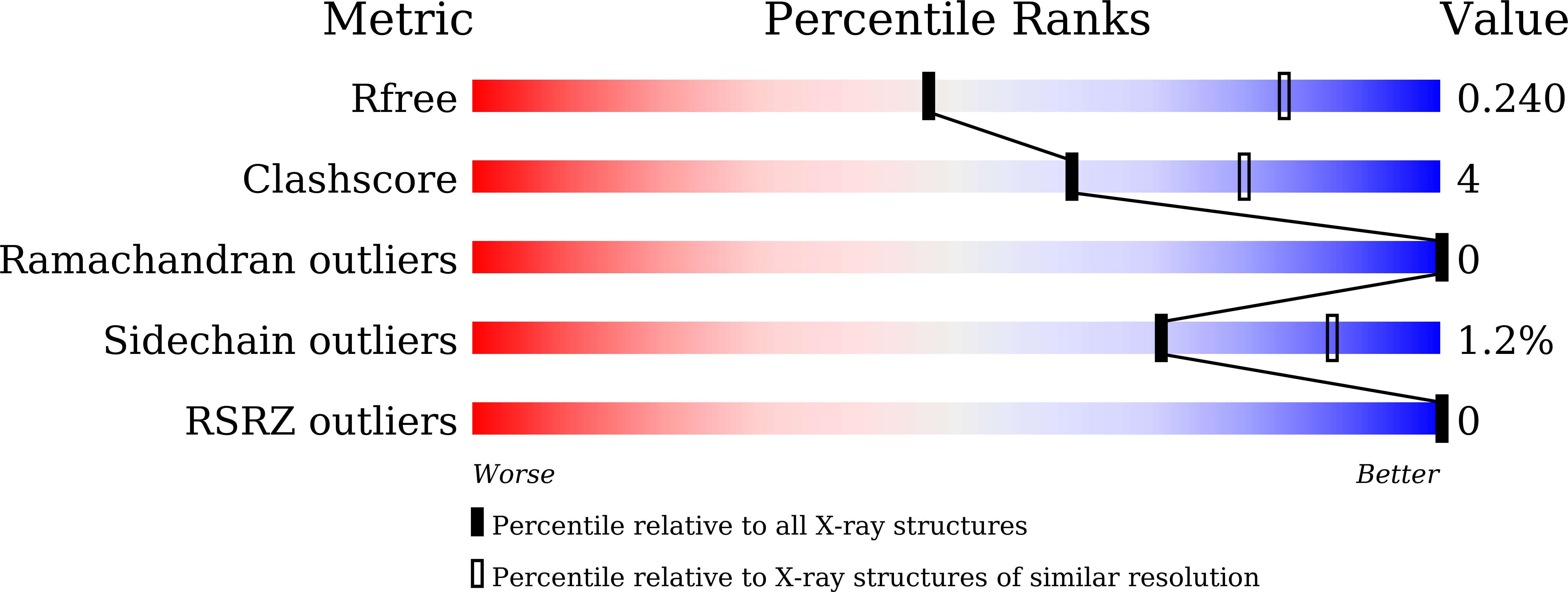
Crystal structure of human acetylcholinesterase in complex with tacrine: Implications for drug discovery
Dileep, K., Ihara, K., Mishima-Tsumagari, C., Kukimoto-Niino, M., Yonemochi, M., Hanada, K., Shirouzu, M., Zhang, K.Y.(2022) Int J Biol Macromol 210: 172-181