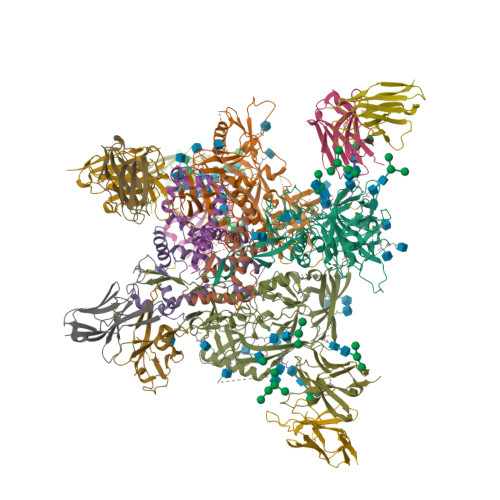
Complementary antibody lineages achieve neutralization breadth in an HIV-1 infected elite neutralizer.
van Schooten, J., Schorcht, A., Farokhi, E., Umotoy, J.C., Gao, H., van den Kerkhof, T.L.G.M., Dorning, J., Rijkhold Meesters, T.G., van der Woude, P., Burger, J.A., Bijl, T., Ghalaiyini, R., Torrents de la Pena, A., Turner, H.L., Labranche, C.C., Stanfield, R.L., Sok, D., Schuitemaker, H., Montefiori, D.C., Burton, D.R., Ozorowski, G., Seaman, M.S., Wilson, I.A., Sanders, R.W., Ward, A.B., van Gils, M.J.(2022) PLoS Pathog 18: e1010945-e1010945
- PubMed: 36395347
- DOI: https://doi.org/10.1371/journal.ppat.1010945
- Primary Citation of Related Structures:
7U5G, 7ZLK - PubMed Abstract:
Broadly neutralizing antibodies (bNAbs) have remarkable breadth and potency against most HIV-1 subtypes and are able to prevent HIV-1 infection in animal models. However, bNAbs are extremely difficult to induce by vaccination. Defining the developmental pathways towards neutralization breadth can assist in the design of strategies to elicit protective bNAb responses by vaccination. Here, HIV-1 envelope glycoproteins (Env)-specific IgG+ B cells were isolated at various time points post infection from an HIV-1 infected elite neutralizer to obtain monoclonal antibodies (mAbs). Multiple antibody lineages were isolated targeting distinct epitopes on Env, including the gp120-gp41 interface, CD4-binding site, silent face and V3 region. The mAbs each neutralized a diverse set of HIV-1 strains from different clades indicating that the patient's remarkable serum breadth and potency might have been the result of a polyclonal mixture rather than a single bNAb lineage. High-resolution cryo-electron microscopy structures of the neutralizing mAbs (NAbs) in complex with an Env trimer generated from the same individual revealed that the NAbs used multiple strategies to neutralize the virus; blocking the receptor binding site, binding to HIV-1 Env N-linked glycans, and disassembly of the trimer. These results show that diverse NAbs can complement each other to achieve a broad and potent neutralizing serum response in HIV-1 infected individuals. Hence, the induction of combinations of moderately broad NAbs might be a viable vaccine strategy to protect against a wide range of circulating HIV-1 viruses.
Organizational Affiliation:
Department of Medical Microbiology, Amsterdam Infection & Immunity Institute, Amsterdam UMC, Location AMC, University of Amsterdam, Amsterdam, The Netherlands.