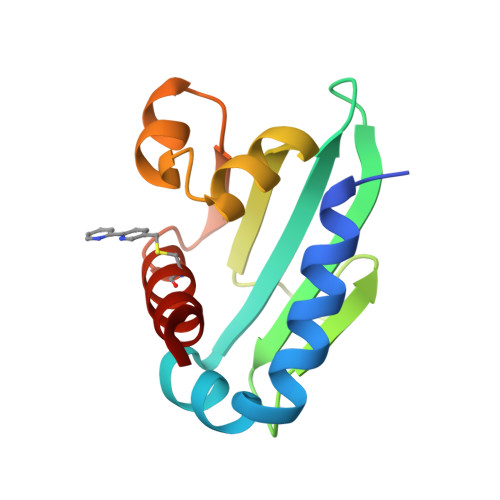
Using BpyAla to generate copper artificial metalloenzymes: a catalytic and structural study.
Klemencic, E., Brewster, R.C., Ali, H.S., Richardson, J.M., Jarvis, A.G.(2024) Catal Sci Technol 14: 1622-1632
- PubMed: 38505507
- DOI: https://doi.org/10.1039/d3cy01648j
- Primary Citation of Related Structures:
8AF2, 8AF3 - PubMed Abstract:
Artificial metalloenzymes (ArMs) have emerged as a promising avenue in the field of biocatalysis, offering new reactivity. However, their design remains challenging due to the limited understanding of their protein dynamics and how the introduced cofactors alter the protein scaffold structure. Here we present the structures and catalytic activity of novel copper ArMs capable of ( R )- or ( S )-stereoselective control, utilizing a steroid carrier protein (SCP) scaffold. To incorporate 2,2'-bipyridine (Bpy) into SCP, two distinct strategies were employed: either Bpy was introduced as an unnatural amino acid (2,2'-bipyridin-5-yl)alanine (BpyAla) using amber stop codon expression or via bioconjugation of bromomethyl-Bpy to cysteine residues. The resulting ArMs proved to be effective at catalysing an enantioselective Friedel-Crafts reaction with SCP_Q111BpyAla achieving the best selectivity with an enantioselectivity of 72% ee ( S ). Interestingly, despite using the same protein scaffold, different attachment strategies for Bpy at the same residue (Q111) led to a switch in the enantiopreference of the ArM. X-ray crystal structures of SCP_Q111CBpy and SCP_Q111BpyAla ArMs with bound Cu(ii) ions unveiled crucial differences in the orientation of the catalytic centre. Combining structural information, alanine scanning studies, and computational analysis shed light on the distinct active sites of the ArMs, clarifying that these active sites stabilise the nucleophilic substrate on different sides of the electrophile leading to the observed switch in enantioselectivity. This work underscores the importance of integrating structural studies with catalytic screening to unravel the intricacies of ArM behaviour and facilitate their development for targeted applications in biocatalysis.
Organizational Affiliation:
EaStCHEM School of Chemistry, University of Edinburgh Joseph Black Building David Brewster Road The King's Buildings Edinburgh EH9 3FJ UK amanda.jarvis@ed.ac.uk.