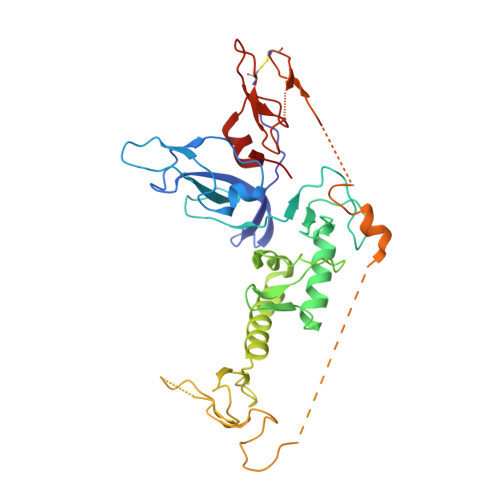
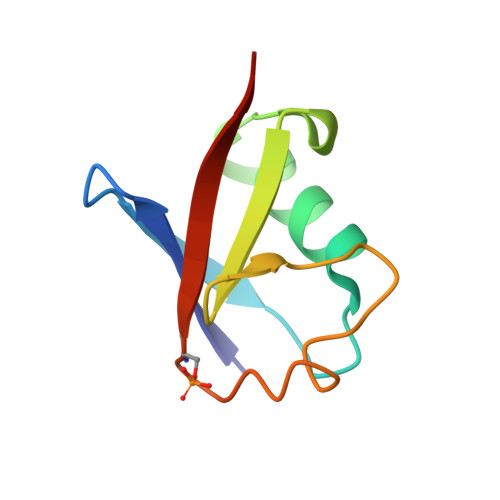
Additional feedforward mechanism of Parkin activation via binding of phospho-UBL and RING0 in trans.
Lenka, D.R., Dahe, S.V., Antico, O., Sahoo, P., Prescott, A.R., Muqit, M.M.K., Kumar, A.(2024) Elife 13
- PubMed: 39221915
- DOI: https://doi.org/10.7554/eLife.96699
- Primary Citation of Related Structures:
8IK6, 8IKM, 8IKT, 8IKV, 8JWV - PubMed Abstract:
Loss-of-function Parkin mutations lead to early-onset of Parkinson's disease. Parkin is an auto-inhibited ubiquitin E3 ligase activated by dual phosphorylation of its ubiquitin-like (Ubl) domain and ubiquitin by the PINK1 kinase. Herein, we demonstrate a competitive binding of the phospho-Ubl and RING2 domains towards the RING0 domain, which regulates Parkin activity. We show that phosphorylated Parkin can complex with native Parkin, leading to the activation of autoinhibited native Parkin in trans . Furthermore, we show that the activator element (ACT) of Parkin is required to maintain the enzyme kinetics, and the removal of ACT slows the enzyme catalysis. We also demonstrate that ACT can activate Parkin in trans but less efficiently than when present in the cis molecule. Furthermore, the crystal structure reveals a donor ubiquitin binding pocket in the linker connecting REP and RING2, which plays a crucial role in Parkin activity.
Organizational Affiliation:
Department of Biological Sciences, Indian Institute of Science Education and Research (IISER) Bhopal, Bhopal, India.