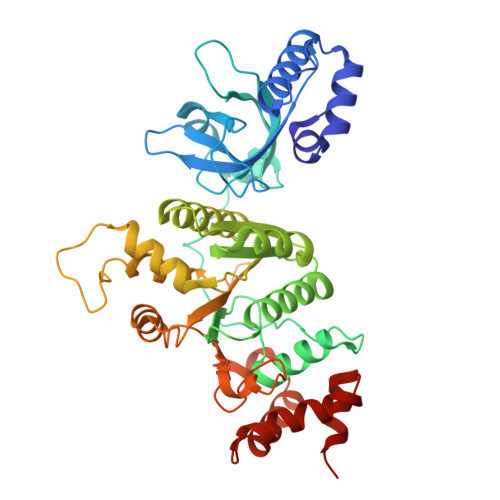
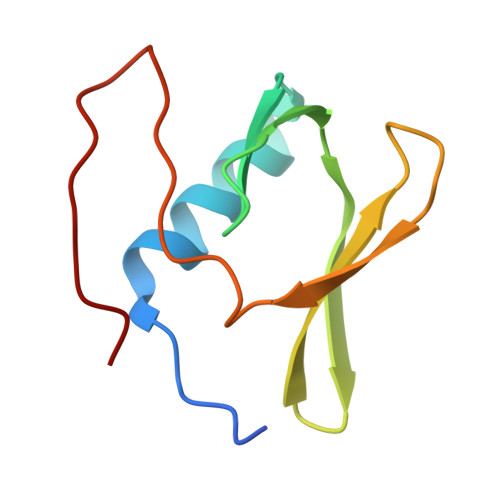
Sm-like protein Rof inhibits transcription termination factor rho by binding site obstruction and conformational insulation.
Said, N., Finazzo, M., Hilal, T., Wang, B., Selinger, T.L., Gjorgjevikj, D., Artsimovitch, I., Wahl, M.C.(2024) Nat Commun 15: 3186-3186
- PubMed: 38622114
- DOI: https://doi.org/10.1038/s41467-024-47439-6
- Primary Citation of Related Structures:
8PTG, 8PTM, 8PTN, 8PTO, 8PTP - PubMed Abstract:
Transcription termination factor ρ is a hexameric, RNA-dependent NTPase that can adopt active closed-ring and inactive open-ring conformations. The Sm-like protein Rof, a homolog of the RNA chaperone Hfq, inhibits ρ-dependent termination in vivo but recapitulation of this activity in vitro has proven difficult and the precise mode of Rof action is presently unknown. Here, our cryo-EM structures of ρ-Rof and ρ-RNA complexes show that Rof undergoes pronounced conformational changes to bind ρ at the protomer interfaces, undercutting ρ conformational dynamics associated with ring closure and occluding extended primary RNA-binding sites that are also part of interfaces between ρ and RNA polymerase. Consistently, Rof impedes ρ ring closure, ρ-RNA interactions and ρ association with transcription elongation complexes. Structure-guided mutagenesis coupled with functional assays confirms that the observed ρ-Rof interface is required for Rof-mediated inhibition of cell growth and ρ-termination in vitro. Bioinformatic analyses reveal that Rof is restricted to Pseudomonadota and that the ρ-Rof interface is conserved. Genomic contexts of rof differ between Enterobacteriaceae and Vibrionaceae, suggesting distinct modes of Rof regulation. We hypothesize that Rof and other cellular anti-terminators silence ρ under diverse, but yet to be identified, stress conditions when unrestrained transcription termination by ρ may be detrimental.
Organizational Affiliation:
Laboratory of Structural Biochemistry, Institute of Chemistry and Biochemistry, Freie Universität Berlin, Takustr. 6, D-14195 Berlin, Germany.