Structural insights into the DNA-binding mechanism of BCL11A: The integral role of ZnF6.
Viennet, T., Yin, M., Jayaraj, A., Kim, W., Sun, Z.J., Fujiwara, Y., Zhang, K., Seruggia, D., Seo, H.S., Dhe-Paganon, S., Orkin, S.H., Arthanari, H.(2024) Structure 32: 2276-2286.e4
- PubMed: 39423807
- DOI: https://doi.org/10.1016/j.str.2024.09.022
- Primary Citation of Related Structures:
8THO, 8TLO - PubMed Abstract:
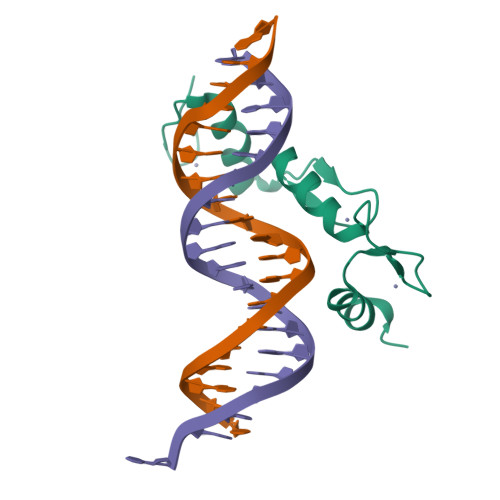
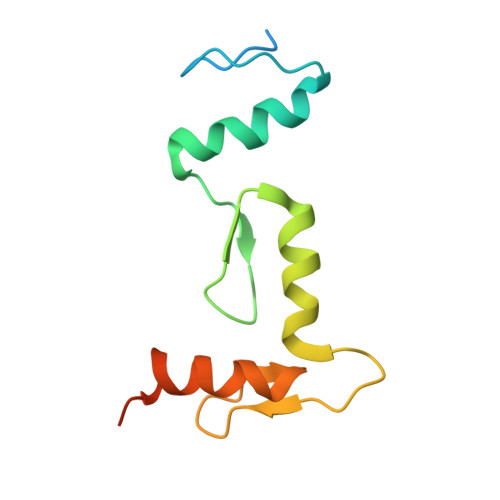
The transcription factor BCL11A is a critical regulator of the switch from fetal hemoglobin (HbF: α 2 γ 2 ) to adult hemoglobin (HbA: α 2 β 2 ) during development. BCL11A binds at a cognate recognition site (TGACCA) in the γ-globin gene promoter and represses its expression. DNA-binding is mediated by a triple zinc finger domain, designated ZnF456. Here, we report comprehensive investigation of ZnF456, leveraging X-ray crystallography and NMR to determine the structures in both the presence and absence of DNA. We delve into the dynamics and mode of interaction with DNA. Moreover, we discovered that the last zinc finger of BCL11A (ZnF6) plays a different role compared to ZnF4 and 5, providing a positive entropic contribution to DNA binding and γ-globin gene repression. Comprehending the DNA binding mechanism of BCL11A opens avenues for the strategic, structure-based design of novel therapeutics targeting sickle cell disease and β-thalassemia.
Organizational Affiliation:
Department of Cancer Biology, Dana-Farber Cancer Institute, Boston, MA, USA; Biological Chemistry and Molecular Pharmacology, Harvard Medical School, Boston, MA, USA.