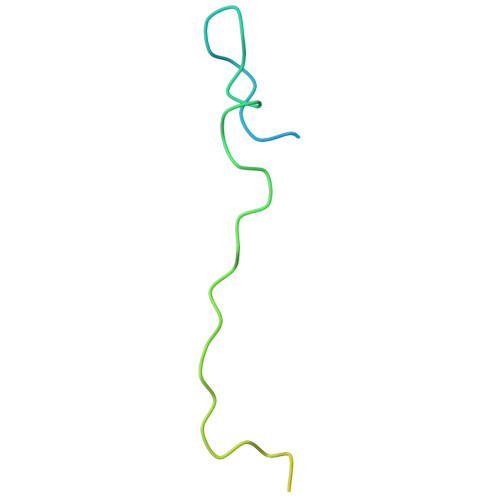
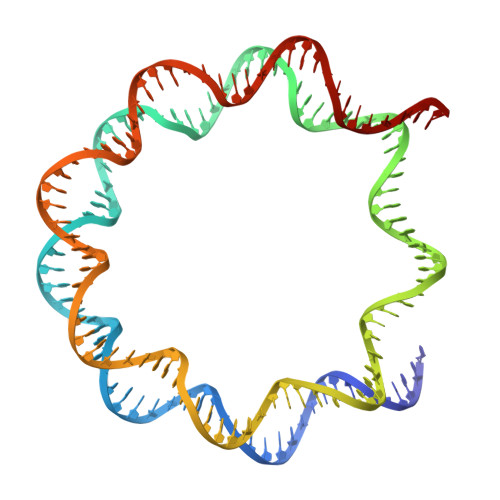
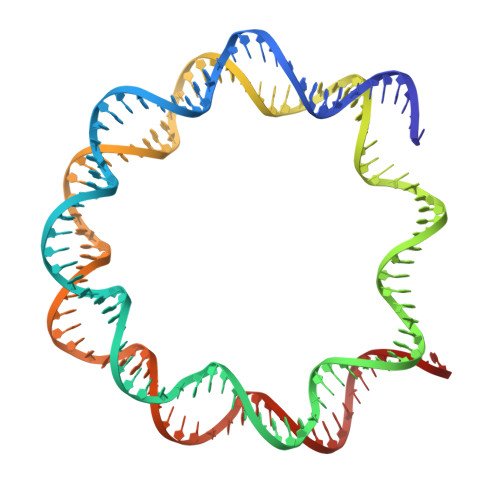
Cancer-associated DNA hypermethylation of Polycomb targets requires DNMT3A dual recognition of histone H2AK119 ubiquitination and the nucleosome acidic patch.
Gretarsson, K.H., Abini-Agbomson, S., Gloor, S.L., Weinberg, D.N., McCuiston, J.L., Kumary, V.U.S., Hickman, A.R., Sahu, V., Lee, R., Xu, X., Lipieta, N., Flashner, S., Adeleke, O.A., Popova, I.K., Taylor, H.F., Noll, K., Windham, C.L., Maryanski, D.N., Venters, B.J., Nakagawa, H., Keogh, M.C., Armache, K.J., Lu, C.(2024) Sci Adv 10: eadp0975-eadp0975
- PubMed: 39196936
- DOI: https://doi.org/10.1126/sciadv.adp0975
- Primary Citation of Related Structures:
8UW1 - PubMed Abstract:
During tumor development, promoter CpG islands that are normally silenced by Polycomb repressive complexes (PRCs) become DNA-hypermethylated. The molecular mechanism by which de novo DNA methyltransferase(s) [DNMT(s)] catalyze CpG methylation at PRC-regulated regions remains unclear. Here, we report a cryo-electron microscopy structure of the DNMT3A long isoform (DNMT3A1) amino-terminal region in complex with a nucleosome carrying PRC1-mediated histone H2A lysine-119 monoubiquitination (H2AK119Ub). We identify regions within the DNMT3A1 amino terminus that bind H2AK119Ub and the nucleosome acidic patch. This bidentate interaction is required for effective DNMT3A1 engagement with H2AK119Ub-modified chromatin in cells. Further, aberrant redistribution of DNMT3A1 to Polycomb target genes recapitulates the cancer-associated DNA hypermethylation signature and inhibits their transcriptional activation during cell differentiation. This effect is rescued by disruption of the DNMT3A1-acidic patch interaction. Together, our analyses reveal a binding interface critical for mediating promoter CpG island DNA hypermethylation, a major molecular hallmark of cancer.
Organizational Affiliation:
Department of Genetics and Development and Herbert Irving Comprehensive Cancer Center, Columbia University Irving Medical Center, New York, NY 10032, USA.