Structural Basis of High-Precision Protein Ligation and Its Application.
Chong, K.H.C., Liu, L., Chua, R., Chai, Y.T., Lu, Z., Liu, R., Tan, E.Y.J., Dong, J., Khoh, Y.H., Lin, J., Zhong, F.L., Lescar, J., Zheng, P., Wu, B.(2025) J Am Chem Soc 147: 1604-1611
- PubMed: 39745918
- DOI: https://doi.org/10.1021/jacs.4c10689
- Primary Citation of Related Structures:
8JTU, 8WKD - PubMed Abstract:
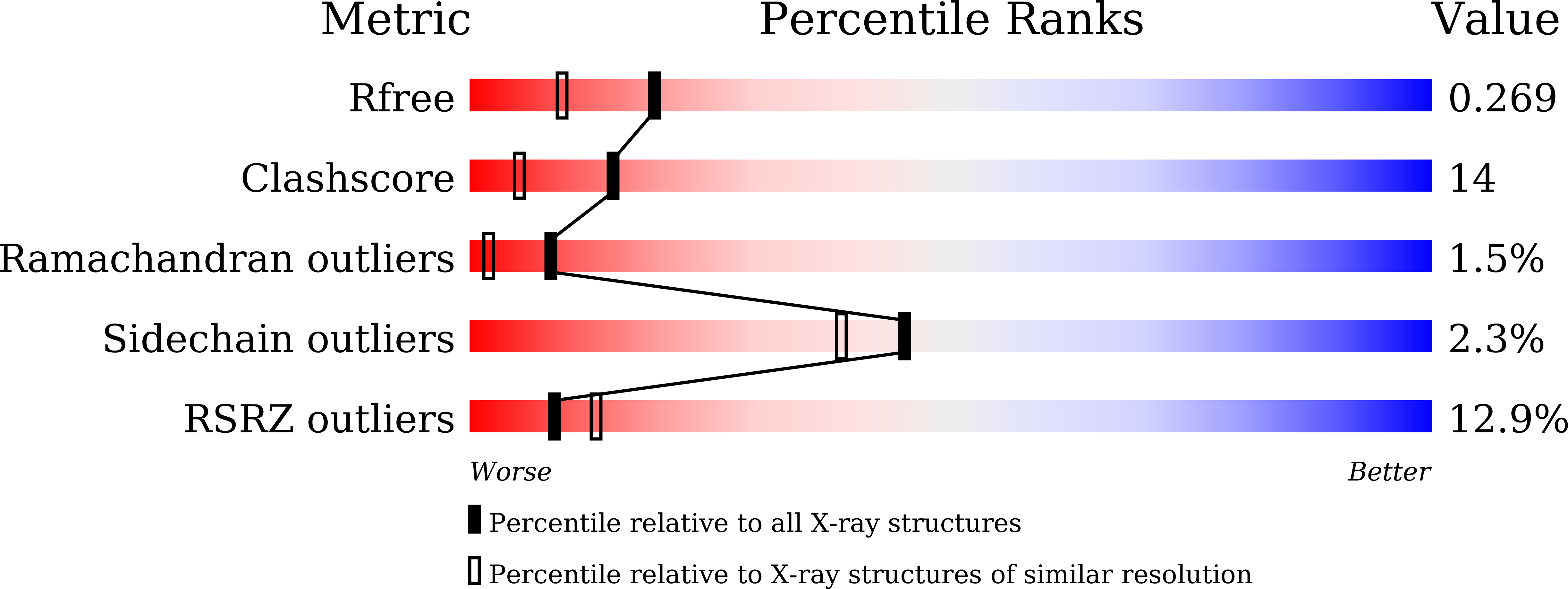
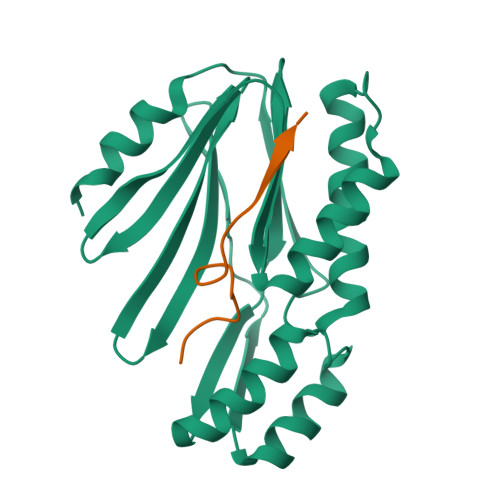
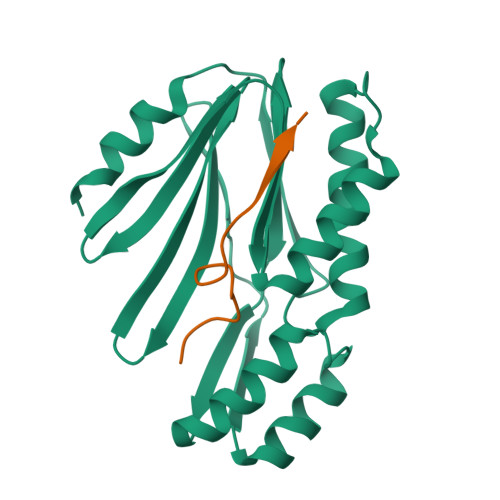
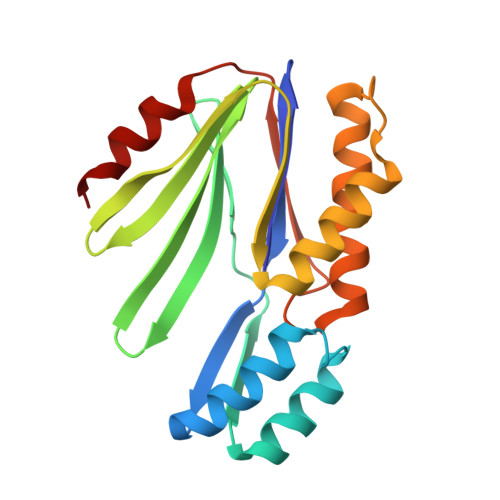
Enzyme-catalyzed protein modifications have become invaluable in diverse applications, outperforming chemical methods in terms of precision, conjugation efficiency, and biological compatibility. Despite significant advances in ligases, such as sortase A and OaAEP1, their use in heterogeneous biological environments remains constrained by limited target sequence specificity. In 2021, Lupas' group introduced Connectase, a family of repurposed archaeal proteases for protein ligations, but its low processivity and lack of structural information have impeded further engineering for practical biological and biophysical applications. Here, we present the X-ray crystallographic structures of MmConnectase ( Methanococcus maripaludis , MmCET) in both apo and substrate-bound forms. Comparative analysis with its inactive paralogue, MjCET ( Methanococcus janaschi ), reveals the structural basis of MmCET's high-precision ligation activity. We propose modifications to the N-terminal substrate recognition motifs to suppress MmCET's reversible protease activity, enabling high-precision protein ligations in complex biological environments, such as serum-containing cell cultures. To further demonstrate the enhanced processivity and precision, single-molecule protein unfolding experiments showed that our optimized Connectase, in conjunction with OaAEP1(C247A), can perform stepwise tandem ligations of protein leading to a well-defined protein polymer.
Organizational Affiliation:
School of Biological Sciences, Nanyang Technological University, 60 Nanyang Drive, Singapore 636921, Singapore.