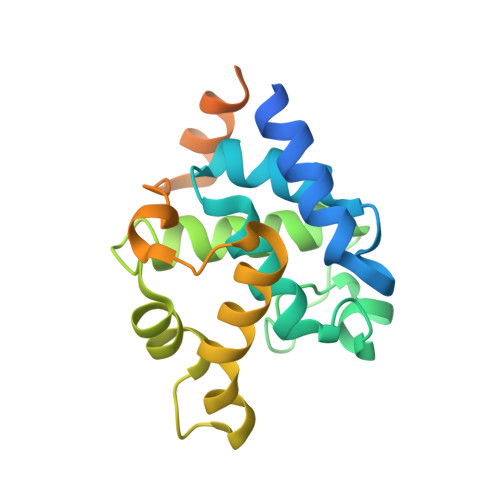
X-ray structure and mutagenesis analyses of Clostridioides difficile endolysin Ecd09610 glucosaminidase domain.
Sekiya, H., Nonaka, Y., Kamitori, S., Miyaji, T., Tamai, E.(2024) Biochem Biophys Res Commun 715: 149957-149957
- PubMed: 38688057
- DOI: https://doi.org/10.1016/j.bbrc.2024.149957
- Primary Citation of Related Structures:
8YXK, 8YXN - PubMed Abstract:
Clostridioides difficile endolysin (Ecd09610) consists of an unknown domain at its N terminus, followed by two catalytic domains, a glucosaminidase domain and endopeptidase domain. X-ray structure and mutagenesis analyses of the Ecd09610 catalytic domain with glucosaminidase activity (Ecd09610CD53) were performed. Ecd09610CD53 was found to possess an α-bundle-like structure with nine helices, which is well conserved among GH73 family enzymes. The mutagenesis analysis based on X-ray structures showed that Glu405 and Asn470 were essential for enzymatic activity. Ecd09610CD53 may adopt a neighboring-group mechanism for a catalytic reaction in which Glu405 acted as an acid/base catalyst and Asn470 helped to stabilize the oxazolinium ion intermediate. Structural comparisons with the newly identified Clostridium perfringens autolysin catalytic domain (AcpCD) in the P1 form and a zymography analysis demonstrated that AcpCD was 15-fold more active than Ecd09610CD53. The strength of the glucosaminidase activity of the GH73 family appears to be dependent on the depth of the substrate-binding groove.
Organizational Affiliation:
Department of Infectious Disease, College of Pharmaceutical Science, Matsuyama University, 4-2 Bunkyo-cho, Matsuyama, Ehime, 790-8578, Japan.