Structural Mechanisms Underlying the Modulation of CXCR4 by Diverse Antagonists.
Sang, X.H., Jiao, H.Z., Huang, Z.W., Hu, H.L.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Soluble cytochrome b562,C-X-C chemokine receptor type 4 | A [auth R] | 486 | Escherichia coli, Homo sapiens | Mutation(s): 0 Gene Names: cybC, CXCR4 | 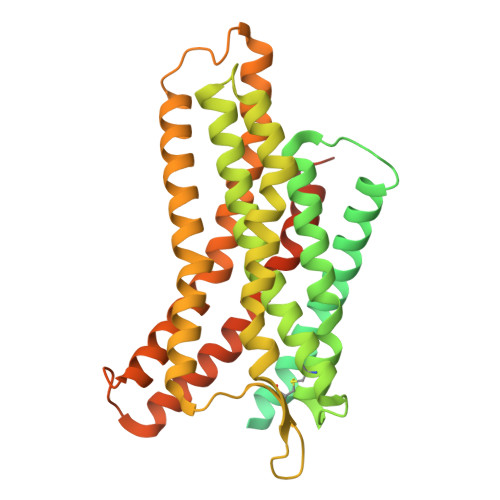 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P61073 (Homo sapiens) Explore P61073 Go to UniProtKB: P61073 | |||||
PHAROS: P61073 GTEx: ENSG00000121966 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P61073 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Nb6 nanobody | B [auth N] | 169 | Lama glama | Mutation(s): 0 | 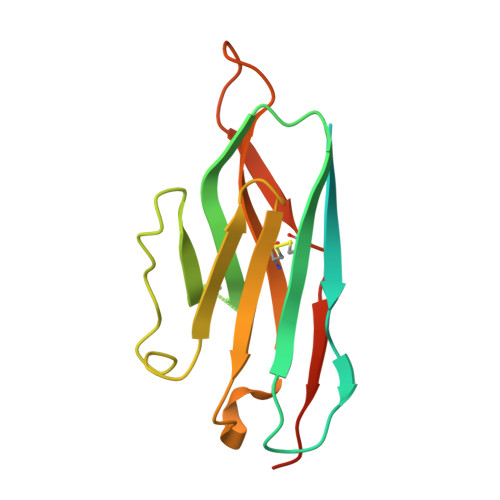 |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| CLR Query on CLR | D [auth R], E [auth R] | CHOLESTEROL C27 H46 O HVYWMOMLDIMFJA-DPAQBDIFSA-N |  | ||
| A1D8L (Subject of Investigation/LOI) Query on A1D8L | C [auth R] | (S)-N1-((1H-Benzo[d]imidazol-2-yl)methyl)-N1-(5,6,7,8-tetrahydroquinolin-8-yl)butane-1,4-diamine C21 H27 N5 WVLHHLRVNDMIAR-IBGZPJMESA-N |  | ||
| Funding Organization | Location | Grant Number |
|---|---|---|
| Other government | China | 32100963 |