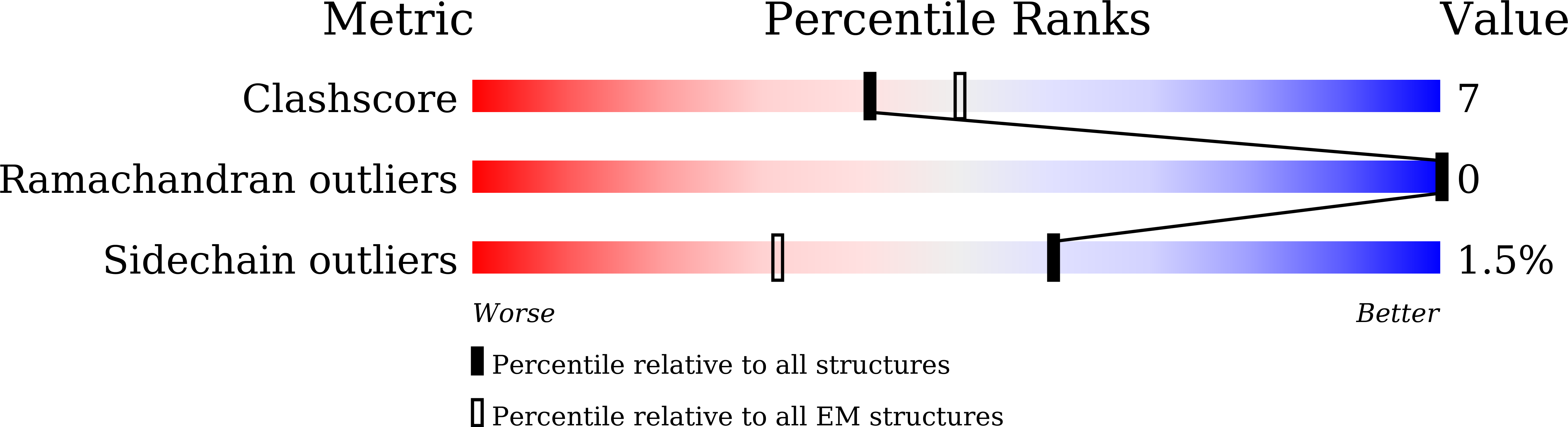Structural mechanisms underlying the modulation of CXCR4 by diverse small-molecule antagonists.
Sang, X., Jiao, H., Meng, Q., Fang, X., Pan, Q., Zhou, J., Qian, T., Zhang, W., Xu, Y., An, J., Huang, Z., Hu, H.(2025) Proc Natl Acad Sci U S A 122: e2425795122-e2425795122
- PubMed: 40063796
- DOI: https://doi.org/10.1073/pnas.2425795122
- Primary Citation of Related Structures:
8ZPL, 8ZPM, 8ZPN - PubMed Abstract:
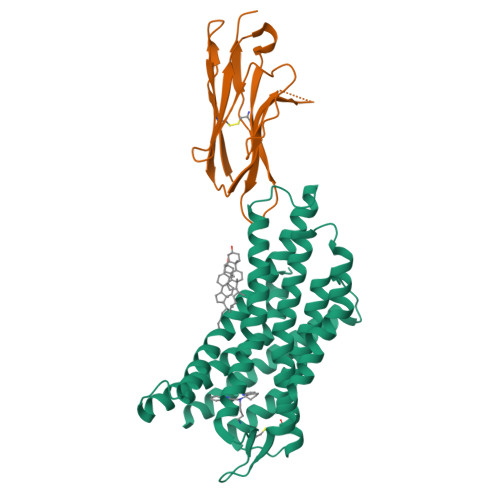
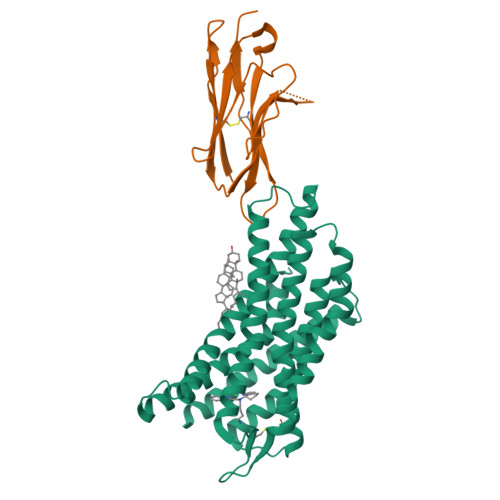
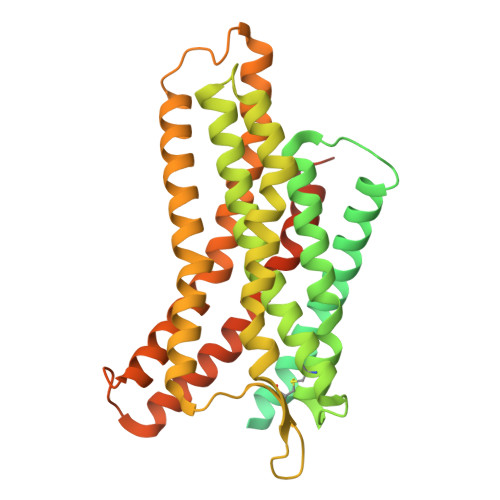
CXCR4 (CXC chemokine receptor type 4), a member of the G protein-coupled receptor superfamily, plays a role in cell migration and functions as a coreceptor for HIV entry. Molecular therapeutics targeting CXCR4 have been under intensive investigation. To date, only two small-molecule antagonist drugs targeting CXCR4, plerixafor (AMD3100) and mavorixafor (AMD070), have been approved. Here, we present the high-resolution structures of CXCR4 complexed with AMD3100 and AMD070, as well as a small-molecule antagonist HF51116 that has very different chemical structure and binding mechanism from AMD3100 and AMD070. The interactions between these antagonists and the receptor are analyzed in details, and the mechanisms of antagonism are elucidated. Both the major and minor subpockets on CXCR4 are found to be involved in binding of these small-molecule antagonists. The distinct conformations of Trp94 2.60 observed in these structures highlight the plasticity of the binding pocket on CXCR4, offering valuable insights into the exploration and refinement of therapeutic strategies targeting this chemokine receptor.
Organizational Affiliation:
Ciechanover Institute of Precision and Regenerative Medicine, School of Medicine, The Chinese University of Hong Kong, Shenzhen 518172, China.