An archaic HLA class I receptor allele diversifies natural killer cell-driven immunity in First Nations peoples of Oceania.
Loh, L., Saunders, P.M., Faoro, C., Font-Porterias, N., Nemat-Gorgani, N., Harrison, G.F., Sadeeq, S., Hensen, L., Wong, S.C., Widjaja, J., Clemens, E.B., Zhu, S., Kichula, K.M., Tao, S., Zhu, F., Montero-Martin, G., Fernandez-Vina, M., Guethlein, L.A., Vivian, J.P., Davies, J., Mentzer, A.J., Oppenheimer, S.J., Pomat, W., Ioannidis, A.G., Barberena-Jonas, C., Moreno-Estrada, A., Miller, A., Parham, P., Rossjohn, J., Tong, S.Y.C., Kedzierska, K., Brooks, A.G., Norman, P.J.(2024) Cell
- PubMed: 39476840
- DOI: https://doi.org/10.1016/j.cell.2024.10.005
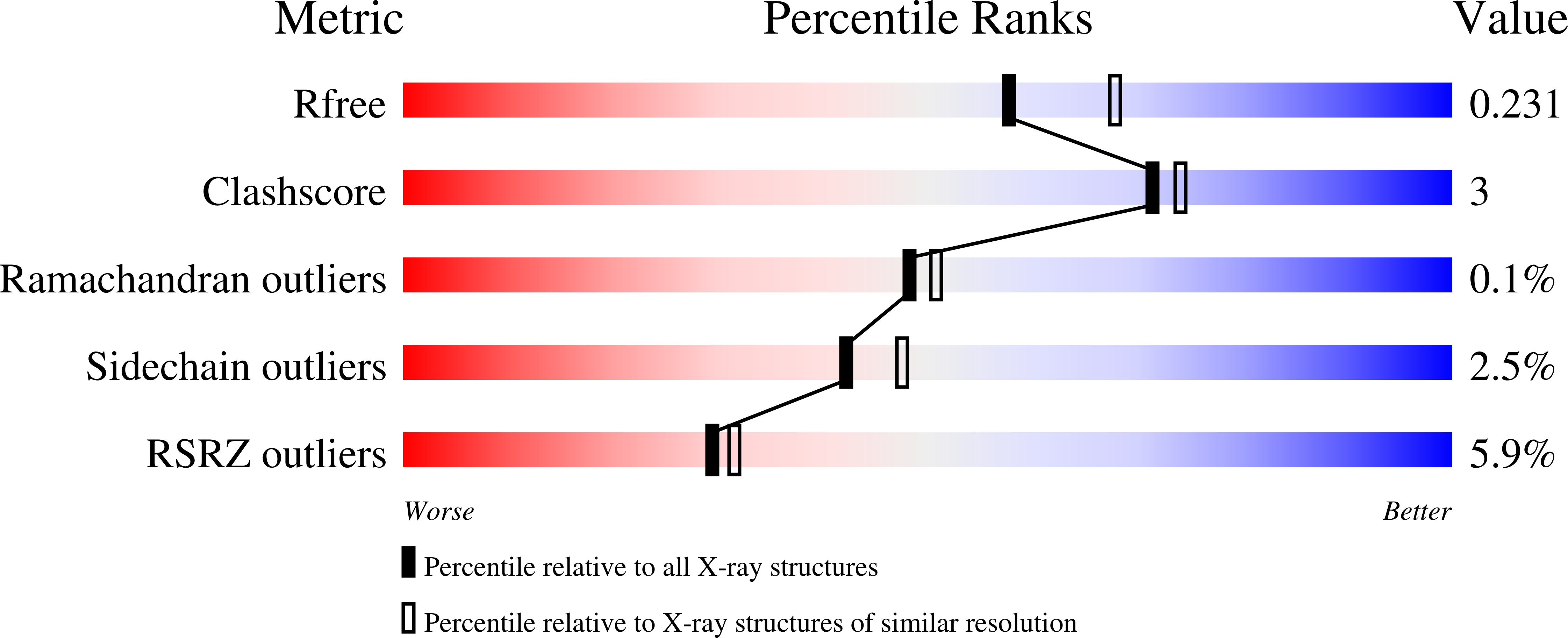
- Primary Citation of Related Structures:
9BL2, 9BL3, 9BL4, 9BL5, 9BL6, 9BL9, 9BLA - PubMed Abstract:
Genetic variation in host immunity impacts the disproportionate burden of infectious diseases that can be experienced by First Nations peoples. Polymorphic human leukocyte antigen (HLA) class I and killer cell immunoglobulin-like receptors (KIRs) are key regulators of natural killer (NK) cells, which mediate early infection control. How this variation impacts their responses across populations is unclear. We show that HLA-A ∗ 24:02 became the dominant ligand for inhibitory KIR3DL1 in First Nations peoples across Oceania, through positive natural selection. We identify KIR3DL1 ∗ 114, widespread across and unique to Oceania, as an allele lineage derived from archaic humans. KIR3DL1 ∗ 114 + NK cells from First Nations Australian donors are inhibited through binding HLA-A ∗ 24:02. The KIR3DL1 ∗ 114 lineage is defined by phenylalanine at residue 166. Structural and binding studies show phenylalanine 166 forms multiple unique contacts with HLA-peptide complexes, increasing both affinity and specificity. Accordingly, assessing immunogenetic variation and the functional implications for immunity are fundamental toward understanding population-based disease associations.
Organizational Affiliation:
Department of Microbiology and Immunology, University of Melbourne at the Peter Doherty Institute for Infection and Immunity, Melbourne, VIC 3000, Australia; Department of Immunology and Microbiology, University of Colorado School of Medicine, Aurora, CO 80045, USA.