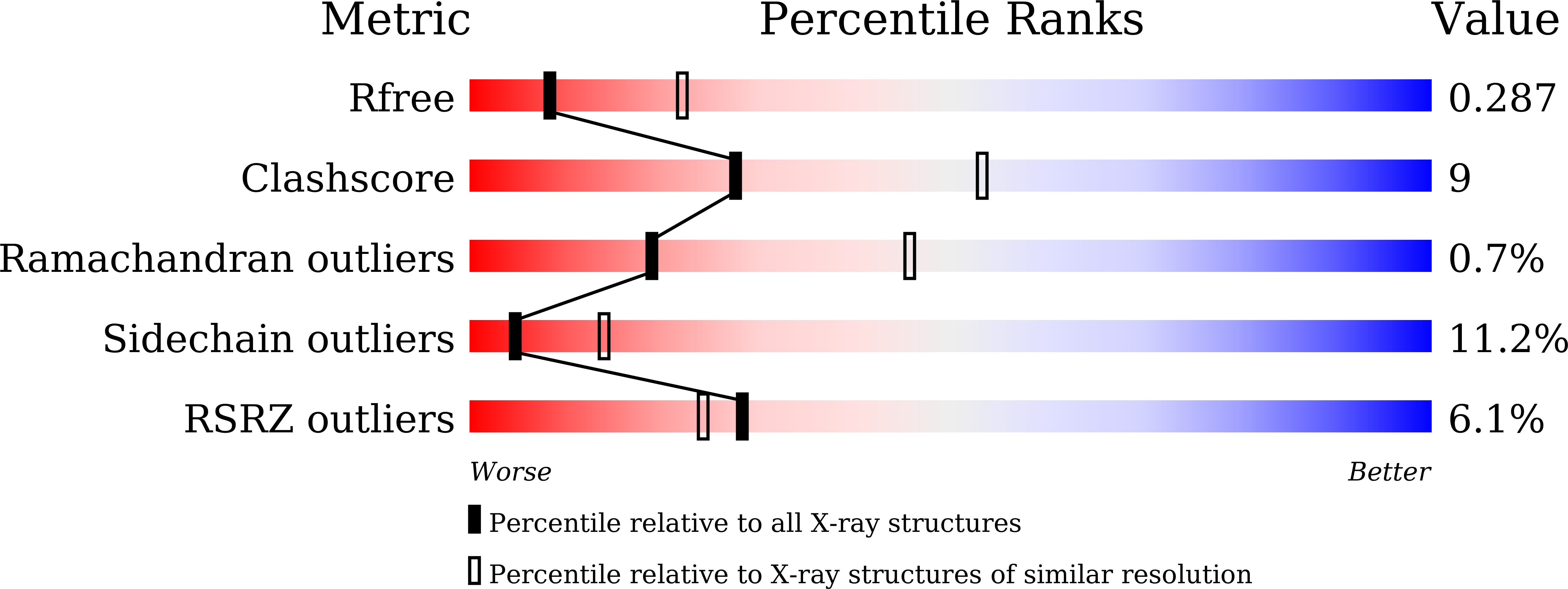
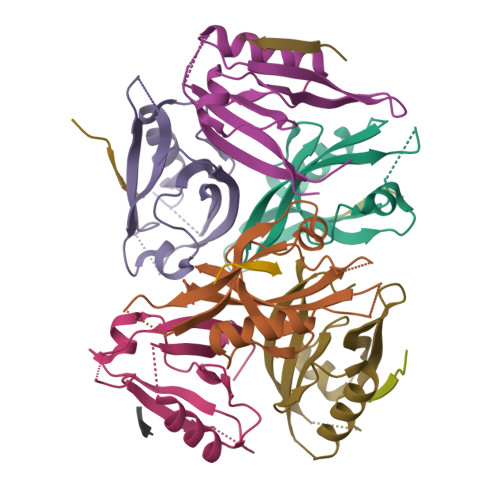
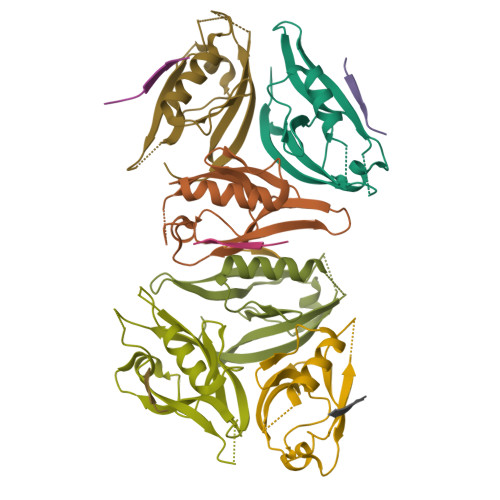
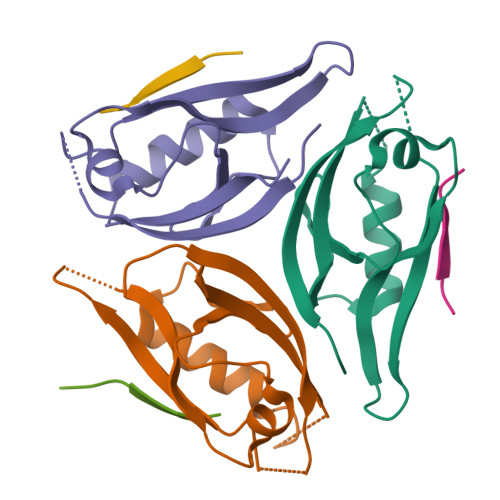
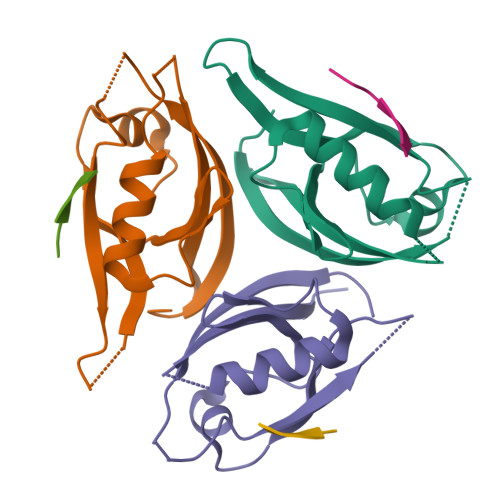
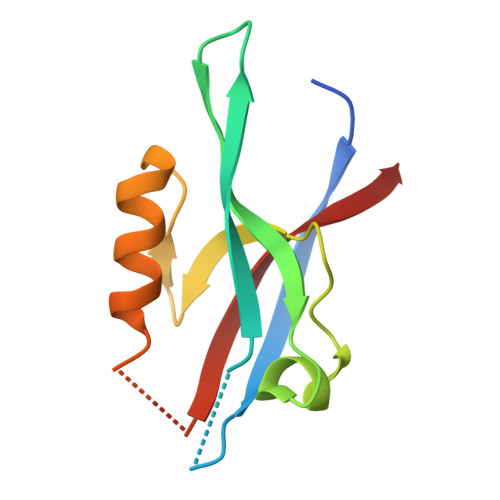
mInsc coordinates Par3 and NuMA condensates for assembly of the spindle orientation machinery in asymmetric cell division.
Huang, S., Fu, M., Gu, A., Zhao, R., Liu, Z., Hua, W., Mao, Y., Wen, W.(2024) Int J Biol Macromol 279: 135126-135126
- PubMed: 39218187
- DOI: https://doi.org/10.1016/j.ijbiomac.2024.135126
- Primary Citation of Related Structures:
9IMP - PubMed Abstract:
As a fundamental process governing the self-renewal and differentiation of stem cells, asymmetric cell division is controlled by several conserved regulators, including the polarity protein Par3 and the microtubule-associated protein NuMA, which orchestrate the assembly and interplay of the Par3/Par6/mInsc/LGN complex at the apical cortex and the LGN/Gαi/NuMA/Dynein complex at the mitotic spindle to ensure asymmetric segregation of cell fate determinants. However, this model, which is well-supported by genetic studies, has been challenged by evidence of competitive interaction between NuMA and mInsc for LGN. Here, the solved crystal structure of the Par3/mInsc complex reveals that mInsc competes with Par6β for Par3, raising questions about how proteins assemble overlapping targets into functional macromolecular complexes. Unanticipatedly, we discover that Par3 can recruit both Par6β and mInsc by forming a dynamic condensate through phase separation. Similarly, the phase-separated NuMA condensate enables the coexistence of competitive NuMA and mInsc with LGN in the same compartment. Bridge by mInsc, Par3/Par6β and LGN/NuMA condensates coacervate, robustly enriching all five proteins both in vitro and within cells. These findings highlight the pivotal role of protein condensates in assembling multi-component signalosomes that incorporate competitive protein-protein interaction pairs, effectively overcoming stoichiometric constraints encountered in conventional protein complexes.
Organizational Affiliation:
Department of Neurosurgery, Huashan Hospital, The Shanghai Key Laboratory of Medical Epigenetics, Institutes of Biomedical Sciences, State Key Laboratory of Medical Neurobiology and MOE Frontiers Center for Brain Science, National Center for Neurological Disorders, School of Basic Medical Sciences, Fudan University, Shanghai 200032, China.