Full Text |
QUERY: Structure Author = "Ayati, M." | MyPDB Login | Search API |
| Search Summary | This query matches 72 Structures. |
Structure Determination MethodologyScientific Name of Source OrganismTaxonomyExperimental MethodPolymer Entity TypeRefinement Resolution (Å)Release DateEnzyme Classification NameSymmetry Type | 1 to 25 of 72 Structures Page 1 of 3 Sort by
The protein complex for DNA replication(2012) Nucleic Acids Res 40: 3208-3217
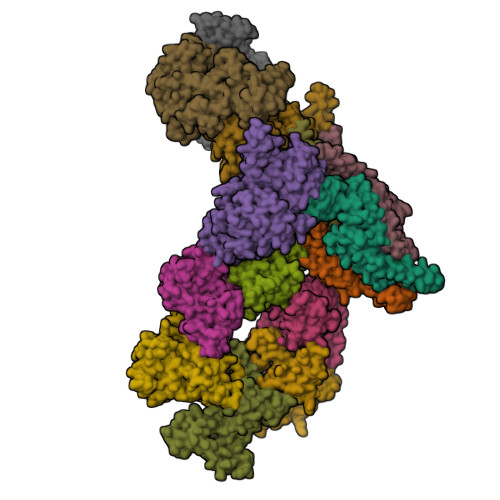
Structure of the eukaryotic replicative CMG helicase and pumpjack motionLi, H., Bai, L., Yuan, Z., Sun, J., Georgescu, R.E., Liu, J., O'Donnell, M.E. (2016) Nat Struct Mol Biol 23: 217-224
Structure of the eukaryotic replicative CMG helicase and pumpjack motionLi, H., Bai, L., Yuan, Z., Sun, J., Georgescu, R.E., Liu, J., O'Donnell, M.E. (2016) Nat Struct Mol Biol 23: 217-224
Structure of the eukaryotic replicative CMG helicase and pumpjack motionLi, H., Bai, L., Yuan, Z., Sun, J., Georgescu, R.E., Liu, J., O'Donnell, M.E. (2016) Nat Struct Mol Biol 23: 217-224
Cryo-EM structure of Mcm2-7 double hexamer on dsDNA(2017) Proc Natl Acad Sci U S A 114: E9529-E9538
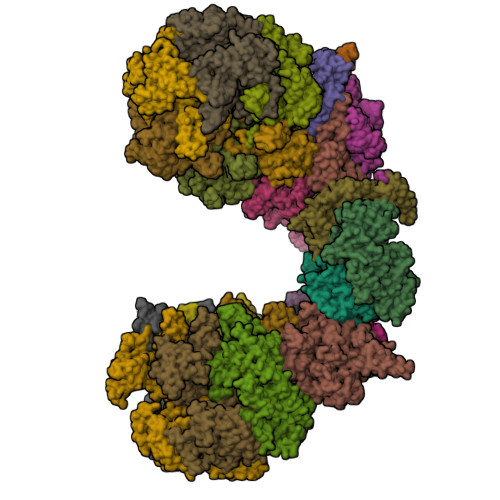
Structure of eukaryotic CMG helicase at a replication forkLi, H., Li, B., Georgescu, R., Yuan, Z., Santos, R., Sun, J., Zhang, D., Yurieva, O., O'Donnell, M.E. (2017) Proc Natl Acad Sci U S A 114: E697-E706
Structure of Eukaryotic CMG Helicase at a Replication Fork and ImplicationsLi, B., Georgescu, R., Yuan, Z., Santos, R., Sun, J., Zhang, D., Yurieva, O., Li, H., O'Donnell, M.E. (2017) Proc Natl Acad Sci U S A 114: E697-E706
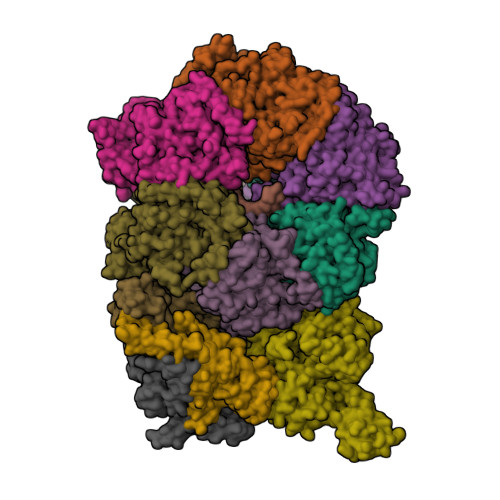
Structural basis of MCM2-7 replicative helicase loading by ORC-Cdc6 and Cdt1Yuan, Z., Riera, A., Bai, L., Sun, J., Spanos, C., Chen, Z.A., Barbon, M., Rappsilber, J., Stillman, B., Speck, C., Li, H. (2017) Nat Struct Mol Biol 24: 316-324
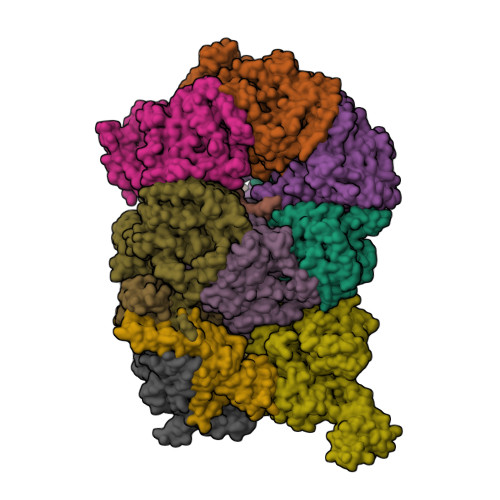
Cryo-EM structure of the Cdt1-MCM2-7 complex in AMPPNP stateZhai, Y., Cheng, E., Wu, H., Li, N., Yung, P.Y., Gao, N., Tye, B.K. (2017) Nat Struct Mol Biol 24: 300-308
Re-refinement of the MCM2-7 double hexamer using ISOLDE(2018) Acta Crystallogr D Struct Biol 74: 519-530
S. cerevisiae MCM double hexamer bound to duplex DNAAbid Ali, F., Pye, V.E., Douglas, M.E., Locke, J., Nans, A., Diffley, J.F.X., Costa, A. (2017) Nat Commun 8: 2241-2241
S. cerevisiae CMG-Pol epsilon-DNAAbid Ali, F., Purkiss, A.G., Cheung, A., Costa, A. (2018) Nat Commun 9: 5061-5061
Structure of Ctf4 trimer in complex with one CMG helicaseYuan, Z., Georgescu, R., Bai, L., Santos, R., Donnell, M., Li, H. (2019) Elife 8:
Structure of Ctf4 trimer in complex with two CMG helicasesYuan, Z., Georgescu, R., Bai, L., Santos, R., Donnell, M., Li, H. (2019) Elife 8:
Structure of Ctf4 trimer in complex with three CMG helicasesYuan, Z., Georgescu, R., Bai, L., Santos, R., Donnell, M., Li, H. (2019) Elife 8:
D. melanogaster CMG-DNA, State 1AEickhoff, P., Martino, F., Costa, A. (2019) Cell Rep 28: 2673-2688.e8
D. melanogaster CMG-DNA, State 1BEickhoff, P., Martino, F., Costa, A. (2019) Cell Rep 28: 2673-2688.e8
D. melanogaster CMG-DNA, State 2AEickhoff, P., Martino, F., Costa, A. (2019) Cell Rep 28: 2673-2688.e8
D. melanogaster CMG-DNA, State 2BEickhoff, P., Martino, F., Costa, A. (2019) Cell Rep 28: 2673-2688.e8
Cryo-EM structure of an MCM loading intermediateMiller, T.C.R., Locke, J., Costa, A. (2019) Nature 575: 704-710
Cryo-EM structure of the CMG Fork Protection Complex at a replication fork - Conformation 1Yeeles, J., Baretic, D., Jenkyn-Bedford, M. (2020) Mol Cell 78: 926-940.e13
Cryo-EM Structure of the Fork Protection Complex Bound to CMG at a Replication Fork - conformation 2 MCM CTD:ssDNAYeeles, J., Baretic, D., Jenkyn-Bedford, M. (2020) Mol Cell 78: 926-940.e13
Atomic model of mutant Mcm2-7 hexamer with Mcm6 WHD truncationYuan, Z., Schneider, S., Dodd, T., Riera, A., Bai, L., Yan, C., Magdalou, I., Ivanov, I., Stillman, B., Li, H., Speck, C. (2020) Proc Natl Acad Sci U S A 117: 17747-17756
Atomic model of pre-insertion mutant OCCM-DNA complex(ORC-Cdc6-Cdt1-Mcm2-7 with Mcm6 WHD truncation)Yuan, Z., Schneider, S., Dodd, T., Riera, A., Bai, L., Yan, C., Magdalou, I., Ivanov, I., Stillman, B., Li, H., Speck, C. (2020) Proc Natl Acad Sci U S A 117: 17747-17756
Atomic model of the mutant OCCM (ORC-Cdc6-Cdt1-Mcm2-7 with Mcm6 WHD truncation) loaded on DNA at 10.5 A resolutionYuan, Z., Schneider, S., Dodd, T., Riera, A., Bai, L., Yan, C., Magdalou, I., Ivanov, I., Stillman, B., Li, H., Speck, C. (2020) Proc Natl Acad Sci U S A 117: 17747-17756
1 to 25 of 72 Structures Page 1 of 3 Sort by |