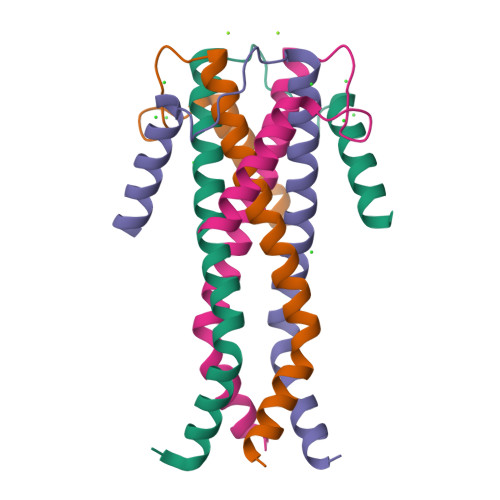
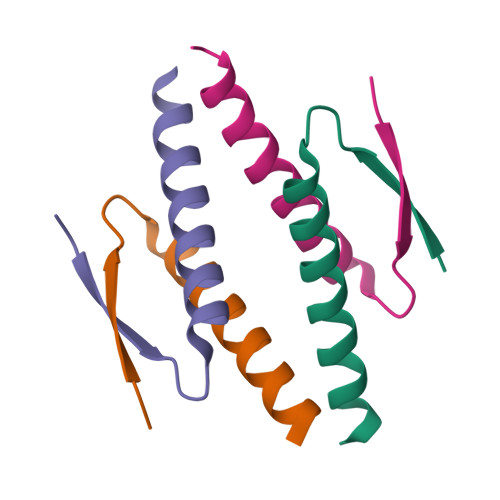
Structure Determination MethodologyScientific Name of Source OrganismRefinement Resolution (Å)Enzyme Classification Name | Cuppari, A., Fernandez-Millan, P., Rubio-Cosials, A., Tarres-Sole, A., Lyonnais, S., Sola, M. (2019) Nucleic Acids Res 47: 6519-6537 | Released | 2019-06-05 | | Method | X-RAY DIFFRACTION 3.05 Å | | Organisms | | | Macromolecule | | | Unique Ligands | 1PE, GOL |
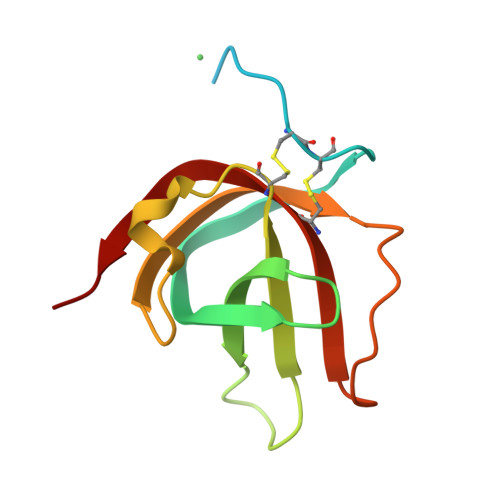
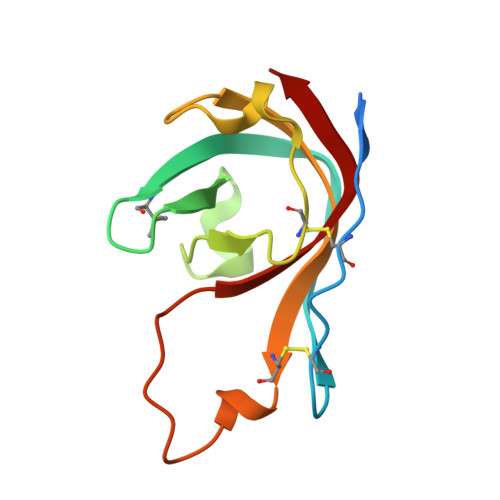
Fernandez-Millan, P., Cuppari, A., Tarres-Sole, A., Rubio-Cosials, A., Lyonnais, S., Sola, M. (2019) Nucleic Acids Res 47: 6519-6537 | Released | 2019-06-05 | | Method | X-RAY DIFFRACTION 3.1 Å | | Organisms | | | Macromolecule | | | Unique Ligands | 1PE, TLA |
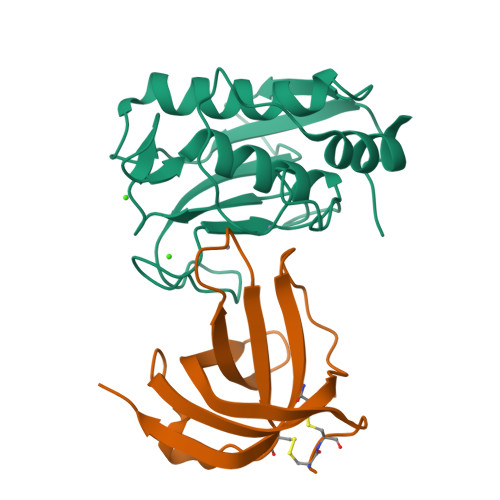
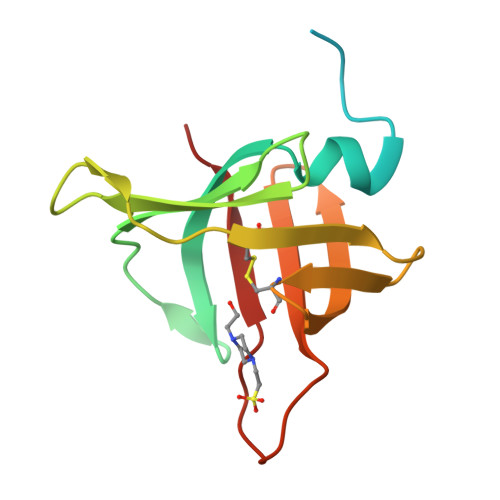
Gomis-Ruth, F.X., Guevara, T., Cuppari, A., Korschgen, H., Schmitz, C., Kuske, M., Yiallouros, I., Floehr, J., Jahnen-Dechent, W., Stocker, W. (2019) Sci Rep 9: 14683-14683 | Released | 2019-10-23 | | Method | X-RAY DIFFRACTION 3 Å | | Organisms | | | Macromolecule | | | Unique Ligands | NAG, ZN | | Unique branched monosaccharides | BMA, FUC, NAG |
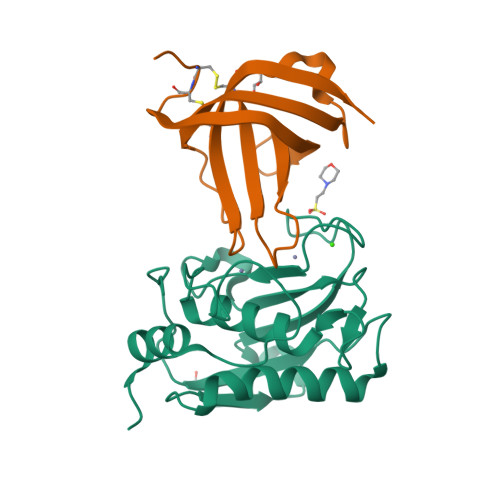
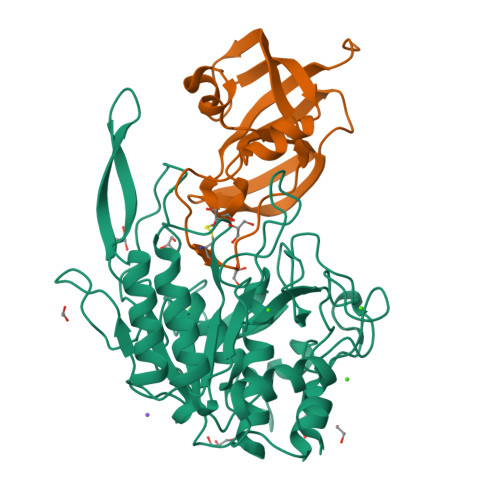
Eckhard, U., Gomis-Ruth, F.X. (2021) Proc Natl Acad Sci U S A 118: | Released | 2021-04-14 | | Method | X-RAY DIFFRACTION 2.8 Å | | Organisms | | | Macromolecule | | | Unique Ligands | NA, NAG, SO4, ZN | | Unique branched monosaccharides | BMA, FUC, GLC, MAN, NAG |
Crespo, I., Boer, D.R. (2022) Comput Struct Biotechnol J 20: 757-765 | Released | 2022-03-09 | | Method | X-RAY DIFFRACTION 1.76 Å | | Organisms | | | Macromolecule | |
Schmitz, C.A., Madej, M., Potempa, J., Sola, M. (2022) Nucleic Acids Res 50: 12558-12577 | Released | 2022-12-14 | | Method | X-RAY DIFFRACTION 2.41 Å | | Organisms | | | Macromolecule | | | Unique Ligands | BEF, FMT, GOL, MG, ZN |
Schmitz, C.A., Madej, M., Potempa, J., Sola, M. (2022) Nucleic Acids Res 50: 12558-12577 | Released | 2022-12-14 | | Method | X-RAY DIFFRACTION 1.91 Å | | Organisms | | | Macromolecule | | | Unique Ligands | BEF, CL, FMT, GOL, MG, ZN |
Schmitz, C.A., Madej, M., Potempa, J., Sola, M. (2022) Nucleic Acids Res 50: 12558-12577 | Released | 2022-12-14 | | Method | X-RAY DIFFRACTION 2.4 Å | | Organisms | | | Macromolecule | | | Unique Ligands | BEF, FMT, GOL, MG, ZN |
Boer, D.R., Crespo, I. (2022) Comput Struct Biotechnol J 20: 757-765 | Released | 2022-03-09 | | Method | X-RAY DIFFRACTION 1.89 Å | | Organisms | | | Macromolecule | | | Unique Ligands | CL, MG |
Potempa, J., Ksiazek, M., Goulas, T., Cuppari, A., Rodriguez-Banqueri, A., Arolas, J.L., Lopez-Pelegrin, M., Garcia-Ferrer, I., Guevara, T. (2023) Chem Sci 14: 869-888 | Released | 2022-12-21 | | Method | X-RAY DIFFRACTION 1.7 Å | | Organisms | | | Macromolecule | | | Unique Ligands | NI |
Potempa, J., Ksiazek, M., Goulas, T., Cuppari, A., Rodriguez-Banqueri, A., Arolas, J.L., Lopez-Pelegrin, M., Garcia-Ferrer, I., Guevara, T. (2023) Chem Sci 14: 869-888 | Released | 2022-12-21 | | Method | X-RAY DIFFRACTION 1.85 Å | | Organisms | | | Macromolecule | | | Unique Ligands | CA, ZN |
Potempa, J., Ksiazek, M., Goulas, T., Cuppari, A., Rodriguez-Banqueri, A., Arolas, J.L., Lopez-Pelegrin, M., Garcia-Ferrer, I., Guevara, T. (2023) Chem Sci 14: 869-888 | Released | 2022-12-21 | | Method | X-RAY DIFFRACTION 1.35 Å | | Organisms | | | Macromolecule | | | Unique Ligands | CA, GOL, MES, ZN |
Gomis-Ruth, F.X. (2023) Chem Sci 14: 869-888 | Released | 2022-12-21 | | Method | X-RAY DIFFRACTION 2.4 Å | | Organisms | | | Macromolecule | | | Unique Ligands | EDO |
Gomis-Ruth, F.X. (2023) Chem Sci 14: 869-888 | Released | 2022-12-21 | | Method | X-RAY DIFFRACTION 2 Å | | Organisms | | | Macromolecule | | | Unique Ligands | GOL |
Gomis-Ruth, F.X. (2023) Chem Sci 14: 869-888 | Released | 2022-12-21 | | Method | X-RAY DIFFRACTION 1.8 Å | | Organisms | | | Macromolecule | | | Unique Ligands | EPE |
Gomis-Ruth, F.X. (2023) Chem Sci 14: 869-888 | Released | 2022-12-21 | | Method | X-RAY DIFFRACTION 1.1 Å | | Organisms | | | Macromolecule | | | Unique Ligands | CA, EDO, GOL, NA |
|