Full Text |
QUERY: Citation Author = "Del, G." | MyPDB Login | Search API |
| Search Summary | This query matches 11 Structures. |
Structure Determination MethodologyScientific Name of Source OrganismTaxonomyExperimental MethodPolymer Entity TypeRefinement Resolution (Å)Release DateMembrane Protein AnnotationSymmetry TypeSCOP Classification | 1 to 11 of 11 Structures Page 1 of 1 Sort by
Crystal structure of acid-sensing ion channel in complex with psalmotoxin 1 at low pH(2012) Nature 489: 400-405
Crystal structure of acid-sensing ion channel in complex with psalmotoxin 1 at high pH(2012) Nature 489: 400-405
Structure of acid-sensing ion channel in complex with snake toxinBaconguis, I., Bohlen, C.J., Goehring, A., Julius, D., Gouaux, E. (2014) Cell 156: 717-729
Structure of acid-sensing ion channel in complex with snake toxin and amilorideBaconguis, I., Bohlen, C.J., Goehring, A., Julius, D., Gouaux, E. (2014) Cell 156: 717-729
Cesium sites in the crystal structure of acid-sensing ion channel in complex with snake toxinBaconguis, I., Bohlen, C.J., Goehring, A., Julius, D., Gouaux, E. (2014) Cell 156: 717-729
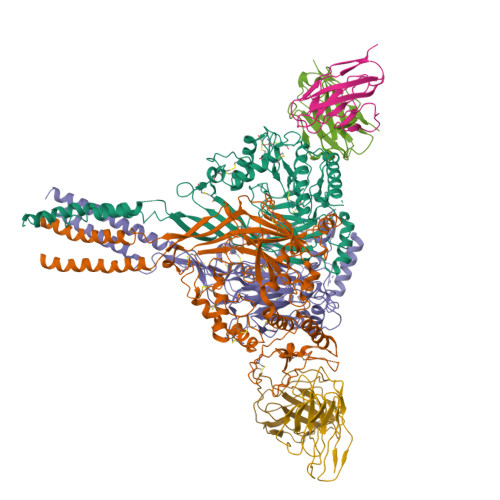
Architecture of fully occupied GluA2 AMPA receptor - TARP complex elucidated by single particle cryo-electron microscopyZhao, Y., Chen, S., Yoshioka, C., Baconguis, I., Gouaux, E. (2016) Nature 536: 108-111
Cryo-EM structure of ENaCNoreng, S., Bharadwaj, A., Posert, R., Yoshioka, C., Baconguis, I. (2018) Elife 7:
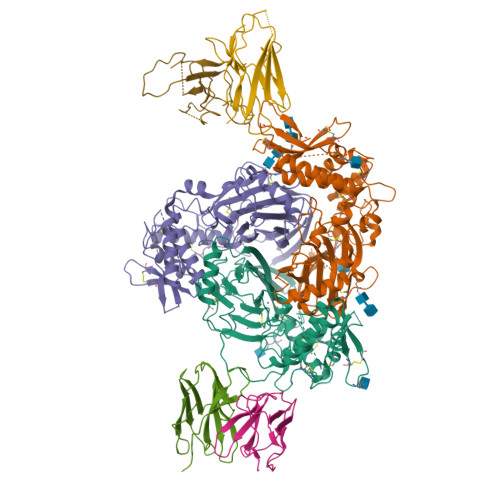
Full-length human ENaC ECDPosert, R., Baconguis, I., Noreng, S., Bharadwaj, A., Houser, A. (2020) Elife 9:
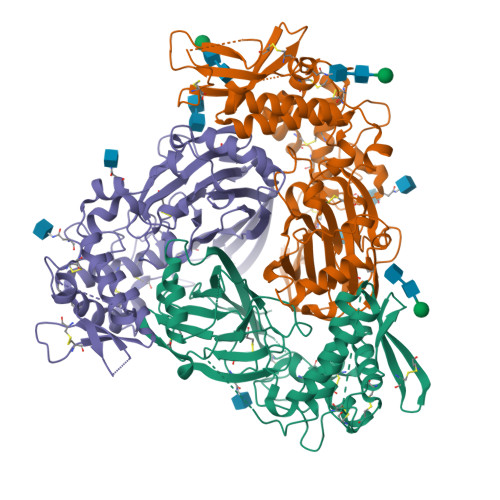
Human SCNN1D-SCNN1B-SCNN1G ENaC trimer(2025) Structure 33: 349-362.e4
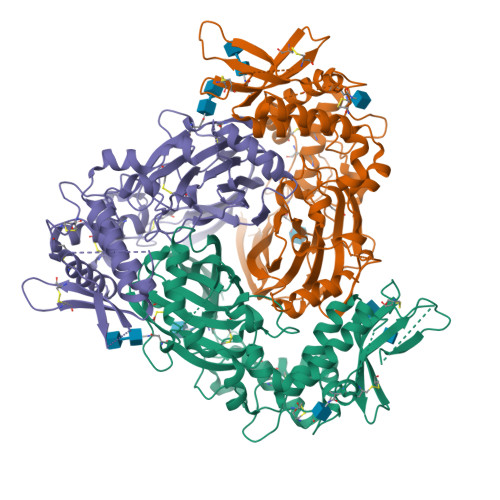
Human SCNN1B-SCNN1B-SCNN1G ENaC trimer(2025) Structure 33: 349-362.e4
Human SCNN1B-SCNN1G ENaC dimers(2025) Structure 33: 349-362.e4
1 to 11 of 11 Structures Page 1 of 1 Sort by |