Full Text |
QUERY: Source Organism Taxonomy Name (Full Lineage) = "Thrixopelma pruriens" | MyPDB Login | Search API |
| Search Summary | This query matches 9 Structures. |
Structure Determination MethodologyScientific Name of Source OrganismTaxonomyExperimental MethodPolymer Entity TypeRefinement Resolution (Å)Release DateMembrane Protein AnnotationSymmetry Type | 1 to 9 of 9 Structures Page 1 of 1 Sort by
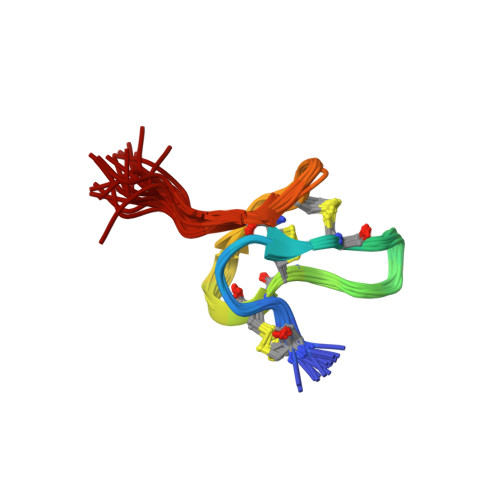
Solution structure of protoxin-1(2014) Curr Biol 24: 473-483
NMR solution structure of ProTx-II(2016) J Biological Chem 291: 17049-17065
Crystal structure of tarantula venom peptide Protoxin-IITabor, A., McCarthy, S., Reyes, F.E. (2017) J Am Chem Soc 139: 13063-13075
NMR solution structure of engineered Protoxin-II analog(2017) Sci Rep 7: 39662-39662
Solution NMR structure of spider toxin analogue [E17K]ProTx-II(2019) ACS Chem Biol 14: 118-130
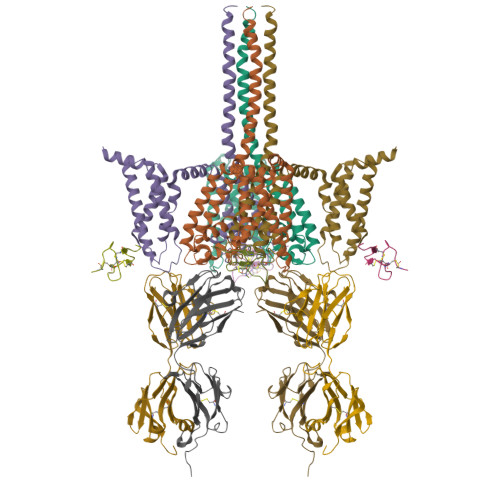
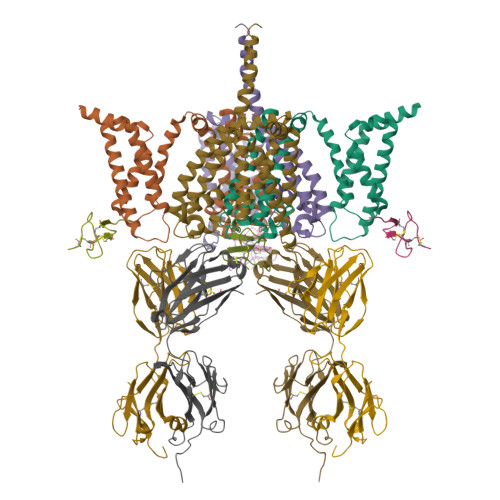
Structural basis of Nav1.7 inhibition by a gating-modifier spider toxinXu, H., Koth, C.M., Payandeh, J. (2019) Cell 176: 702-715.e14
CryoEM structure of Nav1.7 VSD2 (actived state) in complex with the gating modifier toxin ProTx2Xu, H., Rohou, A., Arthur, C.P., Estevez, A., Ciferri, C., Payandeh, J., Koth, C.M. (2019) Cell 176: 702
CryoEM structure of Nav1.7 VSD2 (deactived state) in complex with the gating modifier toxin ProTx2Xu, H., Rohou, A., Arthur, C.P., Estevez, A., Ciferri, C., Payandeh, J., Koth, C.M. (2019) Cell 176: 702
Tarantula venom peptide Protoxin-I bound to full-length human voltage-gated sodium channel 1.8 (NaV1.8)Neumann, B., McCarthy, S., Gonen, S. (2025) Nat Commun 16: 1459-1459
1 to 9 of 9 Structures Page 1 of 1 Sort by |