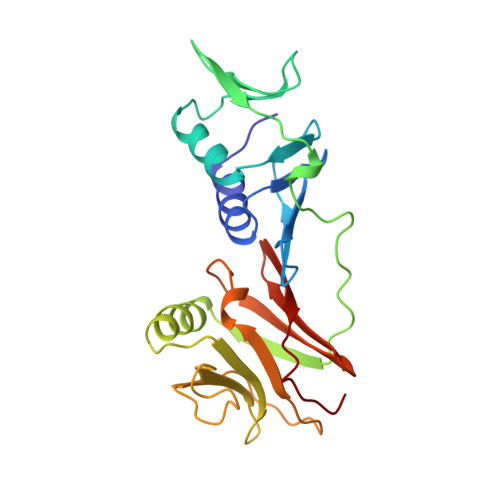
Building a replisome from interacting pieces: sliding clamp complexed to a peptide from DNA polymerase and a polymerase editing complex.
Shamoo, Y., Steitz, T.A.(1999) Cell 99: 155-166
- PubMed: 10535734
- DOI: https://doi.org/10.1016/s0092-8674(00)81647-5
- Primary Citation of Related Structures:
1B77, 1B8H, 1CLQ - PubMed Abstract:
We have solved the crystal structures of the bacteriophage RB69 sliding clamp, its complex with a peptide essential for DNA polymerase interactions, and the DNA polymerase complexed with primer-template DNA. The editing complex structure shows a partially melted duplex DNA exiting from the exonuclease domain at an unexpected angle and significant changes in the protein structure. The clamp complex shows the C-terminal 11 residues of polymerase bound in a hydrophobic pocket, and it allows docking of the editing and clamp structures together. The peptide binds to the sliding clamp at a position identical to that of a replication inhibitor peptide bound to PCNA, suggesting that the replication inhibitor protein p21CIP1 functions by competing with eukaryotic polymerases for the same binding pocket on the clamp.
- Department of Molecular Biophysics and Biochemistry, Yale University, New Haven, Connecticut 06520-8814, USA.
Organizational Affiliation: