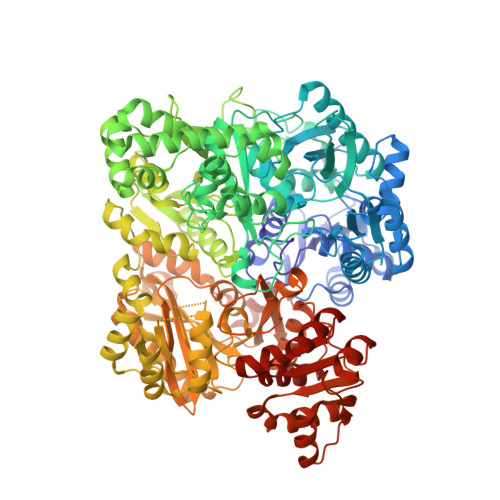
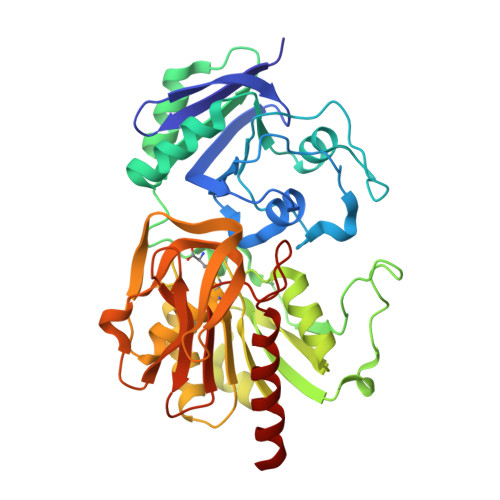
Inactivation of the amidotransferase activity of carbamoyl phosphate synthetase by the antibiotic acivicin.
Miles, B.W., Thoden, J.B., Holden, H.M., Raushel, F.M.(2002) J Biol Chem 277: 4368-4373
- PubMed: 11729189
- DOI: https://doi.org/10.1074/jbc.M108582200
- Primary Citation of Related Structures:
1KEE - PubMed Abstract:
Carbamoyl phosphate synthetase (CPS) from Escherichia coli catalyzes the formation of carbamoyl phosphate from 2 mol of ATP, bicarbonate, and glutamine. CPS was inactivated by the glutamine analog, acivicin. In the presence of ATP and bicarbonate the second-order rate constant for the inactivation of the glutamine-dependent activities was 4.0 x 10(4) m(-1) s(-1). In the absence of ATP and bicarbonate the second-order rate constant for inactivation of CPS was reduced by a factor of 200. The enzyme was protected against inactivation by the inclusion of glutamine in the reaction mixture. The ammonia-dependent activities were unaffected by the incubation of CPS with acivicin. These results are consistent with the covalent labeling of the glutamine-binding site located within the small amidotransferase subunit. The binding of ATP and bicarbonate to the large subunit of CPS must also induce a conformational change within the amidotransferase domain of the small subunit that enhances the nucleophilic character of the thiol group required for glutamine hydrolysis. The acivicin-inhibited enzyme was crystallized, and the three-dimensional structure was determined by x-ray diffraction techniques. The thiol group of Cys-269 was covalently attached to the dihydroisoxazole ring of acivicin with the displacement of a chloride ion.
Organizational Affiliation:
Department of Chemistry, Texas A & M University, College Station, Texas 77842-3012, USA.