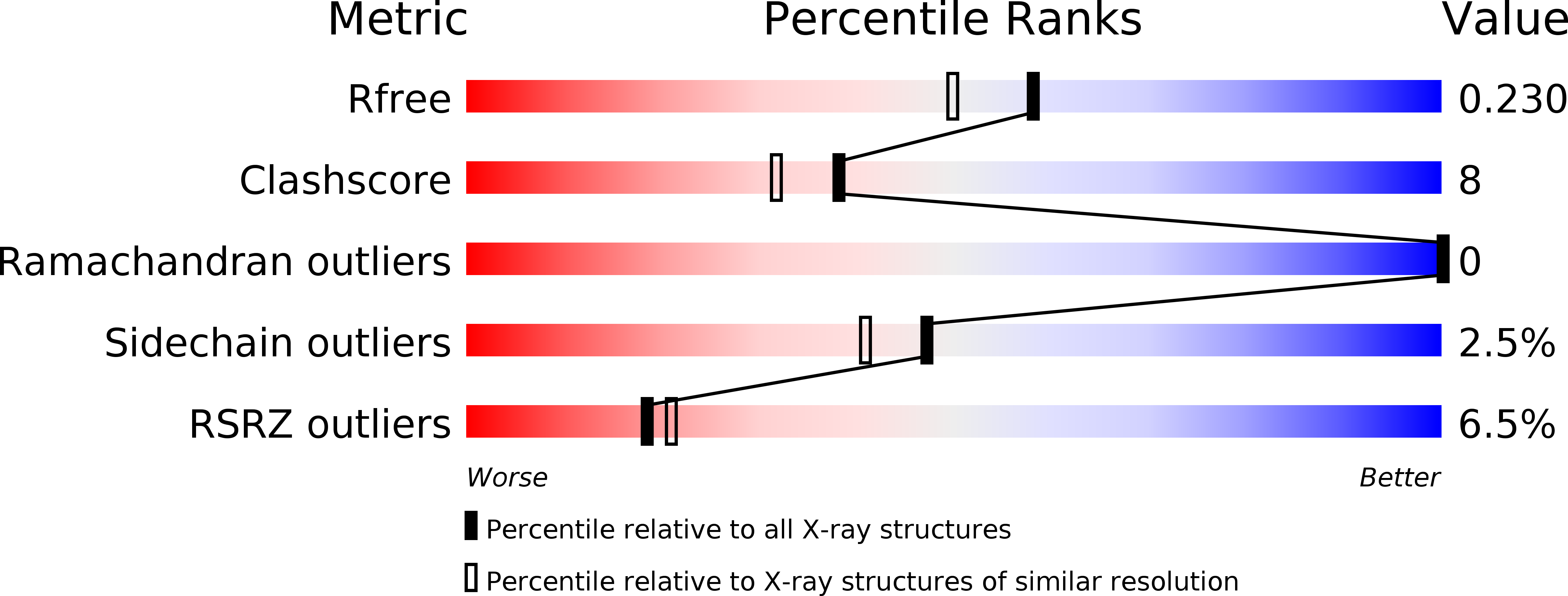
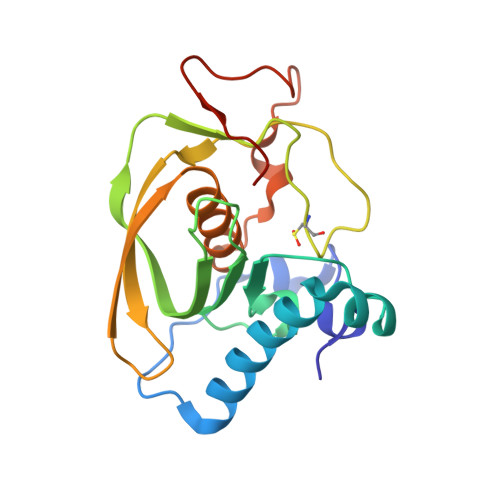
Crystal structure of peptide deformylase from Staphylococcus aureus in complex with actinonin, a naturally occurring antibacterial agent
Yoon, H.J., Kim, H.L., Lee, S.K., Kim, H.W., Kim, H.W., Lee, J.Y., Mikami, B., Suh, S.W.(2004) Proteins 57: 639-642
- PubMed: 15382235
- DOI: https://doi.org/10.1002/prot.20231
- Primary Citation of Related Structures:
1IX1, 1Q1Y
Organizational Affiliation:
Department of Chemistry, College of Natural Sciences, Seoul National University, Seoul, Korea.