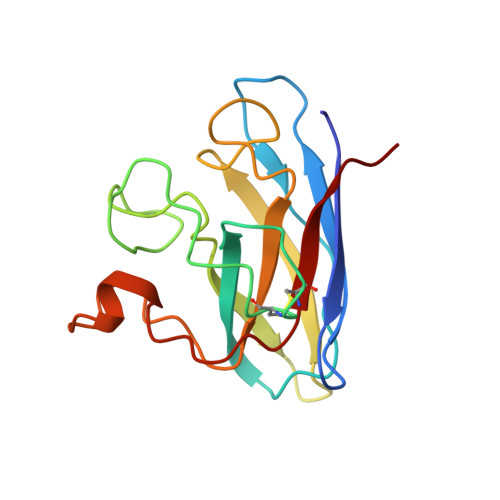
Atomic structures of wild-type and thermostable mutant recombinant human Cu,Zn superoxide dismutase.
Parge, H.E., Hallewell, R.A., Tainer, J.A.(1992) Proc Natl Acad Sci U S A 89: 6109-6113
- PubMed: 1463506
- DOI: https://doi.org/10.1073/pnas.89.13.6109
- Primary Citation of Related Structures:
1SOS - PubMed Abstract:
Superoxide dismutase enzymes protect aerobic organisms from oxygen-mediated free-radical damage. Crystallographic structures of recombinant human Cu,Zn superoxide dismutase have been determined, refined, and analyzed at 2.5 A resolution for wild-type and a designed thermostable double-mutant enzyme (Cys-6----Ala, Cys-111----Ser). The 10 subunits (five dimers) in the crystallographic asymmetric unit form an unusual stable open lattice with 80-A-diameter channels. The 10 independently fit and refined subunits provide high accuracy, error analysis, and insights on loop conformations. There is a helix dipole interaction with the Zn site, and 14 residues form two or more structurally conserved side-chain to main-chain hydrogen bonds that appear critical to active-site architecture, loop conformation, and the increased stability resulting from the Cys-111----Ser mutation.
- Department of Molecular Biology, Scripps Research Institute, La Jolla, CA 92037.
Organizational Affiliation: