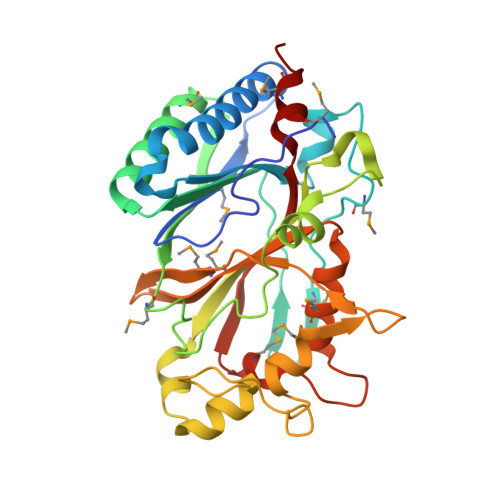
Crystal structures of two novel dye-decolorizing peroxidases reveal a beta-barrel fold with a conserved heme-binding motif.
Zubieta, C., Krishna, S.S., Kapoor, M., Kozbial, P., McMullan, D., Axelrod, H.L., Miller, M.D., Abdubek, P., Ambing, E., Astakhova, T., Carlton, D., Chiu, H.J., Clayton, T., Deller, M.C., Duan, L., Elsliger, M.A., Feuerhelm, J., Grzechnik, S.K., Hale, J., Hampton, E., Han, G.W., Jaroszewski, L., Jin, K.K., Klock, H.E., Knuth, M.W., Kumar, A., Marciano, D., Morse, A.T., Nigoghossian, E., Okach, L., Oommachen, S., Reyes, R., Rife, C.L., Schimmel, P., van den Bedem, H., Weekes, D., White, A., Xu, Q., Hodgson, K.O., Wooley, J., Deacon, A.M., Godzik, A., Lesley, S.A., Wilson, I.A.(2007) Proteins 69: 223-233
- PubMed: 17654545
- DOI: https://doi.org/10.1002/prot.21550
- Primary Citation of Related Structures:
2GVK, 2HAG - PubMed Abstract:
BtDyP from Bacteroides thetaiotaomicron (strain VPI-5482) and TyrA from Shewanella oneidensis are dye-decolorizing peroxidases (DyPs), members of a new family of heme-dependent peroxidases recently identified in fungi and bacteria. Here, we report the crystal structures of BtDyP and TyrA at 1.6 and 2.7 A, respectively. BtDyP assembles into a hexamer, while TyrA assembles into a dimer; the dimerization interface is conserved between the two proteins. Each monomer exhibits a two-domain, alpha+beta ferredoxin-like fold. A site for heme binding was identified computationally, and modeling of a heme into the proposed active site allowed for identification of residues likely to be functionally important. Structural and sequence comparisons with other DyPs demonstrate a conservation of putative heme-binding residues, including an absolutely conserved histidine. Isothermal titration calorimetry experiments confirm heme binding, but with a stoichiometry of 0.3:1 (heme:protein).
- Stanford Synchrotron Radiation Laboratory, Stanford University, Menlo Park, California, USA.
Organizational Affiliation: