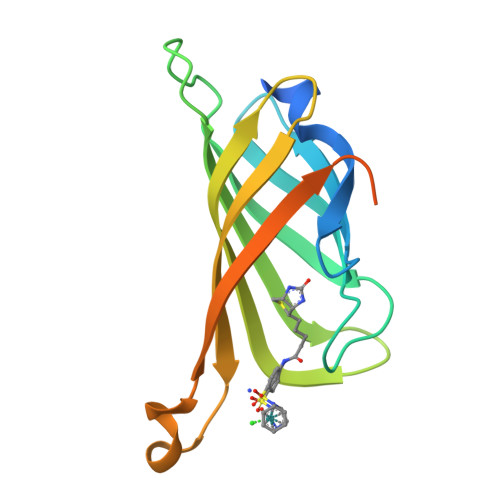
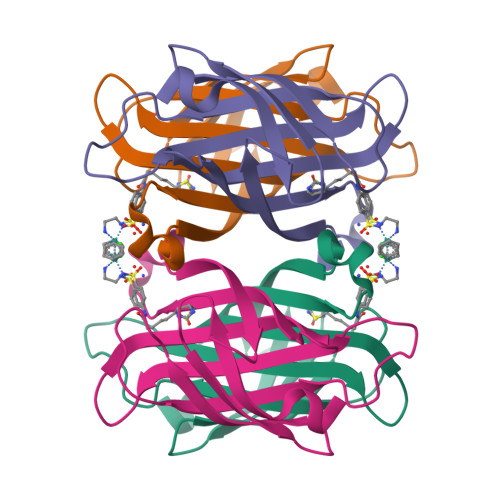
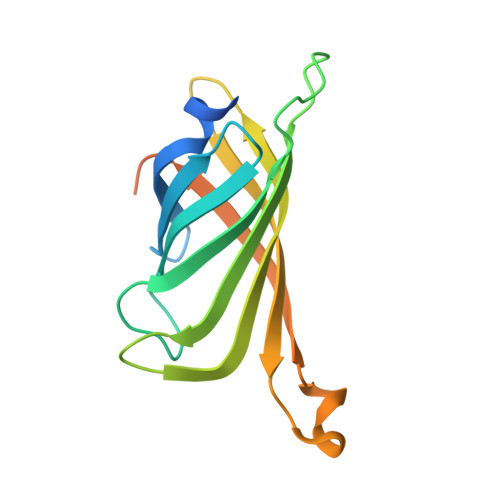
X-ray structure and designed evolution of an artificial transfer hydrogenase
Creus, M., Pordea, A., Rossel, T., Sardo, A., Letondor, C., Ivanova, A., Letrong, I., Stenkamp, R.E., Ward, T.R.(2008) Angew Chem Int Ed Engl 47: 1400-1404
- PubMed: 18176932
- DOI: https://doi.org/10.1002/anie.200704865
- Primary Citation of Related Structures:
2QCB
Organizational Affiliation:
Institute of Chemistry, University of Neuchâtel, Av. Bellevaux 51, CP 158, 2009 Neuchâtel, Switzerland.